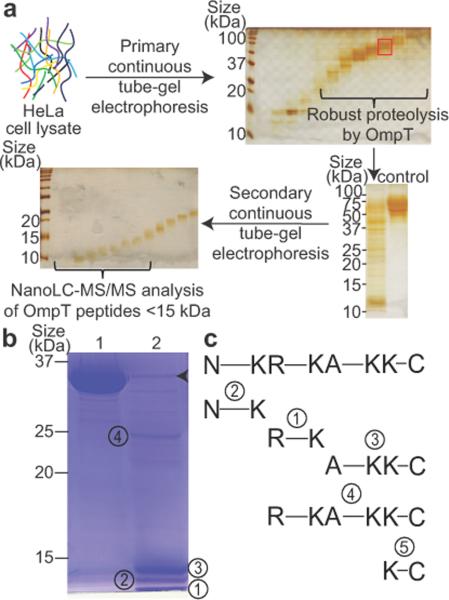
Figure 1.
OmpT-based platform for middle-down proteomics and characterization of OmpT peptides from digestion of a standard protein. (a) The middle-down workflow was illustrated on proteins from a HeLa cell lysate sorted into narrow size ranges by molecular-weight based pre-fractionation (see silver stained gel, top row). A representative OmpT digestion of a fraction containing 50–75 kDa proteins (highlighted in the red box) was visualized by silver staining (left lane, bottom right) along with the control sample with no digestion (right lane). The digested samples were separated further and fractions below ~15 kDa were subjected to nanoLC-MS/MS analysis. (b) Peptide products from digestion of glyceraldehyde 3-phosphate dehydrogenase (GAPDH, 36 kDa) by OmpT were visualized on a Coomassie stained SDS-PAGE gel. Lane 1, GAPDH incubated without OmpT. Lane 2, GAPDH after OmpT digestion. Major peptide products are numbered from 1 through 4. Arrowhead indicates the intact OmpT enzyme. (c) Alignment of identified OmpT peptides by nanoLC-MS/MS with the original GAPDH sequence on top. Peptide cleavage sites are illustrated and N and C represent the protein N and C termini.