Figure 1. In Vivo Assay for ARE-Regulated Translation.
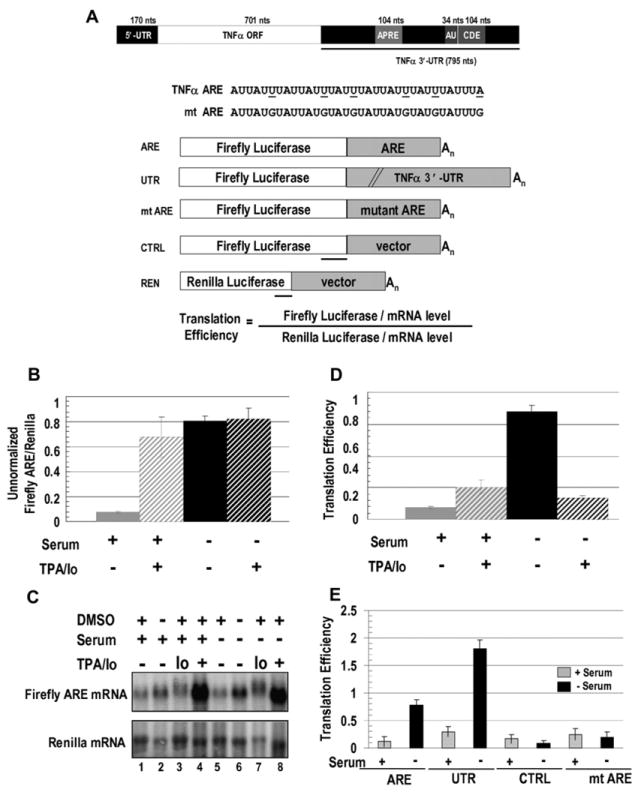
(A) TNFα mRNA with conserved regions, including a splicing regulator (APRE; Osman et al., 1999), the ARE (AU), and a constitutive decay element (CDE; Stoecklin et al., 2003) are shown. Firefly luciferase reporter constructs with the 34 nt TNFα ARE (ARE), the 795 nt TNFα 3′-UTR (UTR), the 34 nt mutant ARE (mt ARE), or a 34 nt vector sequence (CTRL) were cotransfected with a Renilla luciferase reporter (REN). Mutations in mt ARE and regions protected by the probes used in RNase protection assays (RPAs) are underlined.
(B) HEK293 cells were transfected with the ARE and REN reporters and 18 hr later were switched to serum-containing (+) or serum-lacking (−) medium containing either TPA/Io (see Experimental Procedures) or the DMSO solvent. Eighteen hours later, firefly and Renilla luciferase activities were assayed. The values in panels (B), (D), and (E) are averages from at least three transfections ± SD.
(C) Northern blots show ARE- and REN-reporter levels in ± serum, with TPA/Io, Io alone, and/or the solvent DMSO as indicated. Table S2 shows the RNA values.
(D) Luciferase values (B) were normalized to the mRNA levels (C) to obtain translation efficiencies (defined in Figure 1A; see Figure S1 and Tables S1 and S2). Since similar changes upon serum starvation without TPA/Io were observed without normalizing for RNA levels (B), normalization does not artificially produce an apparent increase in translation.
(E) Comparison of translation efficiencies of the ARE and UTR reporters relative to the control reporters, CTRL, and mt ARE in response to the presence or absence of serum without DMSO is shown.
