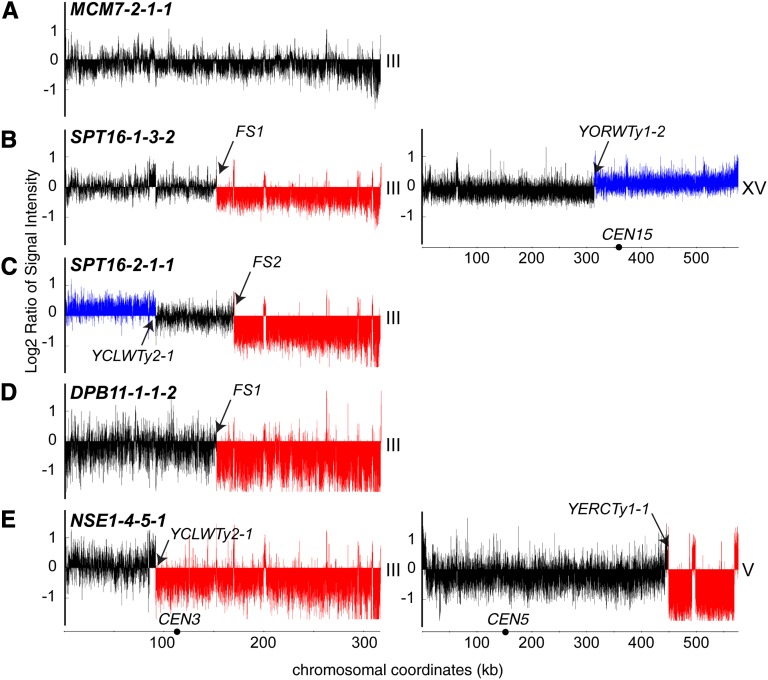
Figure4 .
Comparative genome hybridization microarray analysis of class 3 illegitimate diploids. Genomic DNA was isolated from class 3 (chromosome III arm loss) illegitimate diploids and hybridized to a Saccharomyces cerevisiae whole genome tiling microarray to identify copy number variations. In each histogram, the y-axis represents log2 ratios of probe signal intensities, comparing the indicated strain to a legitimate MATa/α diploid, and the x-axis represents chromosome coordinates. Black arrows indicate breakpoint locations on each chromosome, black circles represent the locations of centromeres, and the chromosome number is indicated to the right of each histogram. A representative histogram for each of the major types of rearrangements observed is shown. (A) Class 3-1 diploid, in which no copy number variation of chromosome III was evident. (B) Class 3-2 diploids have a loss of sequence (red) from the right arm of chromosome III and duplication of sequences (blue) from chromosome XV. (C) Class 3-3 diploids have an amplification of the left arm sequence and a deletion of the right arm sequence of chromosome III. (D) Class 3-4 diploids have a loss of sequence from the right arm of chromosome III without copy number variation on nonhomologous chromosomes. (E) Class 3-5 diploids exhibit a loss of sequence from the right arm of chromosome III and loss of sequence from the right arm of chromosome V.