Abstract
Availability of key nutrients, such as sugars, amino acids, and nitrogen compounds, dictates the developmental programs and the growth rates of yeast cells. A number of overlapping signaling networks—those centered on Ras/protein kinase A, AMP-activated kinase, and target of rapamycin complex I, for instance—inform cells on nutrient availability and influence the cells’ transcriptional, translational, posttranslational, and metabolic profiles as well as their developmental decisions. Here I review our current understanding of the structures of the networks responsible for assessing the quantity and quality of carbon and nitrogen sources. I review how these signaling pathways impinge on transcriptional, metabolic, and developmental programs to optimize survival of cells under different environmental conditions. I highlight the profound knowledge we have gained on the structure of these signaling networks but also emphasize the limits of our current understanding of the dynamics of these signaling networks. Moreover, the conservation of these pathways has allowed us to extrapolate our finding with yeast to address issues of lifespan, cancer metabolism, and growth control in more complex organisms.
YEAST cells finely tune their growth and behavior in accordance with available nutrients. They can adjust their growth rate in response to their nutritional environment by altering the length of their cell cycle over at least a 10-fold range (Brauer et al. 2008). They can adapt to nutritional depletion by engaging one of a number of alternative developmental programs depending on the particular nutritional circumstances. These programs can range from rapid mitotic growth in rich media, to filamentous growth allowing foraging under limiting nutrient conditions, to various distinct quiescent states that reversibly shut down the cell in response to starvation for a single nutrient, to the extreme state of biological stasis following sporulation upon severe starvation.
In metazoans, in which cells are continuously bathed in a uniform sea of nutrients, regulation of metabolic activity, cell growth, or developmental progression at the cellular level is dictated by growth factors, hormones, and modulators. For yeast, nutrients supply not only the substrates for growth but also the signals for growth. That is, nutrients serve not only as the resources by which the cell increases mass and generates energy to propel its biosynthetic activity but also as the signals dictating the metabolic, transcriptional, and developmental programs that optimize survival under the particular nutritional state in which the cell finds itself. Thus, understanding nutrient regulation in yeast requires understanding the dual role of nutrients as metabolites and as signaling molecules and appreciating how those two roles are interconnected.
In this review I describe our current understanding of how the yeast Saccharomyces responds to the two major classes of nutrients, carbon and nitrogen. I will focus on the means by which yeast cells perceive the amount and quality of these classes of nutrients and how they use that information, both singly and in combination, to alter their cellular, metabolic, transcriptional, and developmental landscapes. Other chapters in this series address the means by which Saccharomyces responds to other nutrient classes, including phosphate, sulfur, and amino acids. Moreover, other chapters address the metabolic flow in the cell as well as the various developmental programs yeast can engage. Finally, several excellent reviews have recently appeared that have addressed glucose-induced signaling (Schuller 2003; Johnston and Kim 2005; Santangelo 2006), nitrogen regulation (Magasanik and Kaiser 2002; De Virgilio and Loewith 2006a; De Virgilio and Lowith 2006b), nutrient sensing in fungi (Bahn et al. 2007), and the response of Saccharomyces to starvation (Smets et al. 2010; De Virgilio 2011). Many of the details of topics covered in this chapter, particularly with regard to earlier studies, are elaborated in a recent review (Zaman et al. 2008).
Nutrient Sensing Pathways
Regulatory networks responsive to carbon sources
Yeast cells grow on a wide variety of compounds as sources of energy and as carbon-containing precursors of anabolic metabolism and biomass accumulation (Johnston and Carlson 1992). However, yeast cells consume glucose or fructose in preference to other mono-, di-, and trisaccharides, such as sucrose, raffinose, or trehalose, and prefer any fermentable carbon source over any source, such as glycerol, ethanol, or acetate, that has to be catabolized by oxidative phosphorylation. This hierarchical pattern of consumption is established by allosteric regulation of various key enzymes in glycolysis and gluconeogenesis, described below, and by an extensive transcriptional regulatory network in which glucose represses transcription of genes required for initial catabolism of less favorable sugars and of genes encoding components of the electron transport chain and other mitochrondrial proteins. This latter regulatory process precludes metabolism by oxidative phosphorylation of any nonfermentable carbon sources in the presence of glucose.
Glucose repression of mitochrondrial function is the basis of the Crabtree effect, whereby Saccharomyces ferments glucose to produce ethanol even under aerobic conditions. The Crabtree effect distinguishes Saccharomyces from closely related yeasts such as Kluyveromyces, for example, which do not perform aerobic fermentation. Such fermentation from glucose to ethanol, which yields 2 ATP molecules per molecule of glucose, is much less efficient in energy production than funneling pyruvate, the primary product of glycolysis, into the tricarboxylic acid cycle, which optimally can yield 32 molecules of ATP from each glucose molecule. Aerobic fermentation to ethanol is particularly energetically unfavorable for Saccharomyces since the subsequent introduction into the TCA cycle of the ethanol produced by fermentation requires ATP consumption. Thus, at first glance, aerobic fermentation would appear to be maladaptive.
Several explanations have been proposed to account for aerobic fermentation in Saccharomyces. One hypothesis holds that Saccharomyces cells, which are relatively resistant to ethanol toxicity, may generate ethanol to defend its niche from competing microorganisms in its normal ecological setting of rotting fruit (Thomson et al. 2005). A second explanation is that growth by fermentation minimizes the production of reactive oxygen species that could increase incorporation of mutagenic errors during DNA replication. This explanation has been invoked to explain the presence of metabolic cycles by which Saccharomyces cells promote a burst of fermentation and suppress oxidative phosphorylation during DNA replication even in cells growing on a nonfermentable carbon source (Chen et al. 2007; Silverman et al. 2010). Finally, the Crabtree effect bears striking resemblance to the Warburg effect observed in a variety of cancer cells, a process in which cells consume more glucose than can be funneled through the tricarboxylic acid (TCA) cycle and shunt the excess metabolized glucose into lactate, even under aerobic conditions. A recent hypothesis proposed to account for the Warburg effect is that this energy-inefficient process may actually be quite efficient in producing both reducing potential and anabolic precursors, namely acetyl-CoA, required for the biosynthetic capacity necessary for producing macromolecular components of a new cell. In this model, aerobic fermentation serves as a means of accelerating rapid growth by facilitating mass accumulation (Vander Heiden et al. 2009). Moreover, the model suggests that Saccharomyces in rich medium subscribes to the same exigencies as cancer cells—the need to produce as many progeny in as short a period of time as possible—and aerobic fermentation fulfills that exigency in both settings. Further studies will be required to determine which, if any, of these explanations account for the Crabtree effect and its restriction to the Saccharomyces clade of yeast species (Pfeiffer et al. 2001).
Reflecting the hierarchical pattern of carbon source utilization in yeast, addition of glucose to cells growing on a nonfermentable carbon source results in rapid and sweeping changes in the phosphorylation profile of yeast proteins and the pattern of yeast gene transcripts. Phosphorylation changes occur on a variety of metabolic and cell-cycle–associated proteins as well as a number of transcription factors and chromatin modifiers, consistent with metabolic, proliferative, and transcriptional reprogramming of the cell in response to carbon source changes. More than 40% of genes change their transcript levels by more than two fold within minutes of a shift of cells from glycerol to glucose (Wang et al. 2004; Kresnowati et al. 2006; Zaman et al. 2009). This transition results in activation of genes required for mass accumulation, such as ribosomal protein and ribosomal biogenesis genes, and repression of genes associated with stress response or required for use of alternate carbon sources. A similarly widespread transcriptional reprogramming occurs following depletion of glucose in cells growing on rich medium and the ensuing transition to growth on ethanol (Derisi et al. 1997; Young et al. 2003; Brauer et al. 2005).
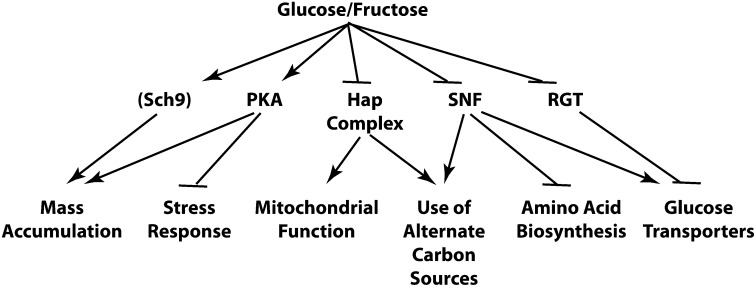
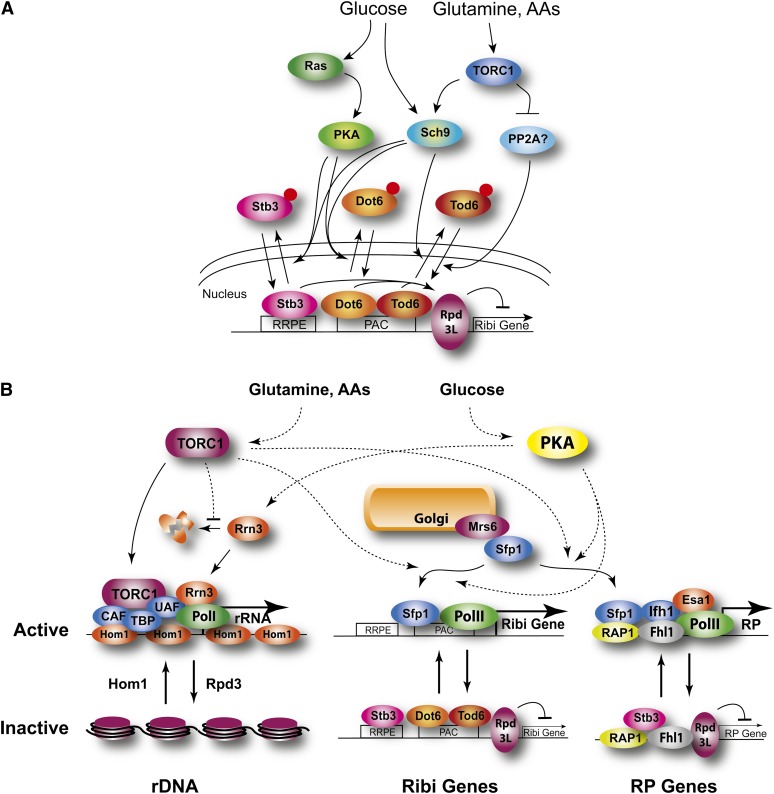
A variety of signaling networks mediate this reprogramming of the metabolic, proliferative, and transcriptional capacity of cells (Figure 1). Different pathways appear to be associated with different processes responsive to the quality and amount of carbon source. For instance, glucose effects on biosynthetic capacity and stress responses are mediated by the protein kinase A pathway, while repression of genes involved in use of alternative carbon sources are mediated predominantly by Snf1 and tuning of the glucose uptake machinery to match glucose levels is effected through the Rgt/Snf3 circuit.
Figure 1 .
An overview of glucose signaling pathways. Different signaling networks respond to availability of fermentable sugars and regulate distinct, albeit overlapping, functions that optimize growth under the particular nutrient status of the cell. Sch9 appears to respond directly to sugar availability but the mechanistic connection is not well defined.
The Ras/protein kinase A pathway:
Most of the glucose-induced signaling in yeast cells proceeds through the Ras/protein kinase A (PKA) pathway (Figure 2). Ninety percent of the transcriptional changes that occur on addition of glucose- to glycerol-grown cells can be recapitulated simply by activating this pathway. Similarly, blocking signaling through the pathway concurrent with glucose addition eliminates most, albeit not all, of the responses. Thus, the PKA pathway is both necessary and sufficient for a majority of the transcriptional responses of the cell to glucose (Zaman et al. 2009). As discussed below, the targets for PKA extend well beyond those involved in transcription and indicate that the kinase exerts effects on growth and development at a variety of levels.
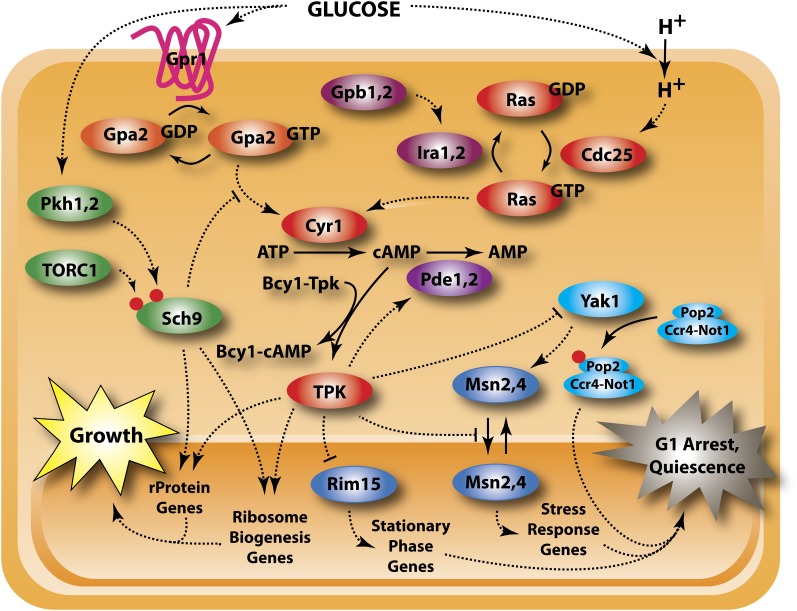
Figure 2 .
The Ras/PKA and Gpa2 pathways. The Ras/PKA pathway plays a central role in regulating growth vs. quiescence in response to the quality and quantity of the available carbon source, primarily by stimulating mass accumulation and inhibiting the stress response. The major input proceeds through Ras, likely in response to glucose-stimulated intracellular acidification, with minor input through the G-protein coupled receptor, Gpr1. Sch9 also mediates the cell growth response to glucose but the indicated link is only speculative. Red dots signify phosphorylation.
Yeast protein kinase A is a heterotetramer comprising two identical regulatory subunits, encoded by BCY1, and two catalytic subunits, encoded by three related genes, TPK1, TPK2, and TPK3. To a first approximation, the catalytic subunits are redundant: any one of the three is sufficient to maintain viability, whereas loss of all three is lethal. However, more nuanced studies suggest that these proteins have somewhat different activities and functional specificities (Robertson and Fink 1998; Ptacek et al. 2005). For instance, Tpk2 appears to stimulate pseudohyphal growth, whereas Tpk3 and Tpk1 inhibit it (Robertson and Fink 1998; Malcher et al. 2011). Moreover, in vitro analysis of the three different subunits indicated overlapping but substantially distinct substrate specificities (Ptacek et al. 2005). The relevance of the in vitro studies to in vivo specificities has not been explored and most genetic and genomic studies highlight the redundancy of the proteins.
cAMP regulates PKA activity by binding to Bcy1, alleviating its inhibitory activity on the catalytic subunits. Cellular levels of cAMP are determined by the competing activities of synthesis from ATP via adenylyl cyclase, encoded by CYR1, and degradation to AMP by low-affinity and high-affinity phosphodiesterases, encoded by PDE1 and PDE2, respectively. Early studies on cAMP levels in cells containing low level constitutive activity of PKA provide compelling evidence that PKA exerts a strong negative feedback on cAMP levels and implicated Pde’s in that feedback (Nikawa et al. 1987). Moreover, Pde1 can be phosphorylated by PKA in vitro and following a glucose pulse in vivo (Ma et al. 1999). However, a decrease in phosphodiesterase activity has never been demonstrated in response to an increase in PKA activity. Nonetheless, the down modulation of Pde activity in response to elevated PKA activity is an essential component of any model that attempts to account for the observed spike in cAMP concentration observed following glucose addition to cells (Williamson et al. 2009). Thus, such down-modulation most likely occurs but through a more subtle mechanism, such as alteration in protein interaction or localization.
Adenylyl cyclase activity is stimulated by two families of small GTP-binding proteins. The yeast homologs of the mammalian ras protooncogene, Ras1 and Ras2, make direct contact with adenylyl cyclase to stimulate its catalytic activity. The Ras proteins cycle between a GDP- and GTP-bound form; only the GTP-bound form activates adenylyl cyclase. The ratio of GTP- to GDP-bound Ras derives from a balance of competing reactions: the guanine nucleotide exchange factor, Cdc25, catalyzes GTP loading of the protein and a GTPase activity intrinsic to the Ras proteins converts the bound GTP to GDP. This intrinsic GTPase can be dramatically enhanced by the activity of two GTPase activating proteins (GAPs), Ira1 and Ira2. Glucose addition to yeast cells elicits a rapid spike in cAMP levels that depends on Ras and mirrors the increase in intracellular levels of GTP-bound Ras (Colombo et al. 1998). Accordingly, glucose could exert its effect by stimulating exchange activity catalyzed by Cdc25 or by inhibiting the GAP activity of Ira proteins. Despite extensive investigations, the mechanism by which glucose affects Ras GTP levels remains unresolved. Ras proteins, like their mammalian counterparts, undergo extensive post-translational modification—including C-terminal proteolytic cleavage, farnesylation, palmitoylation, and carboxymethylation—and are deposited on the inner surface of the plasma membrane by a specialized transport mechanism (Dong et al. 2003; Wang and Deschenes 2006). Addition of glucose to cells elicits a rapid acidification of the yeast cytoplasm (Dechant et al. 2010). While no direct evidence has emerged to show that changes in membrane potential or cytoplasmic acidification affects adenylyl cyclase activity, these are currently the most compelling models for activation of the PKA pathway.
Gpa2, a member of the Gα component of the heterotrimeric G-protein family, also participates in activation of PKA through stimulation of adenylyl cyclase activity. Bacterially expressed Gpa2 bound to GTPγS but not to GDP can associate with yeast adenylyl cyclase in vitro (Peeters et al. 2006). Moreover, induction of the activated GPA2Q300L allele, which encodes a mutant protein defective in GTP hydrolysis and thus remains bound to GTP in vivo, results in the same reconfiguration of the transcriptional profile of cells as does induction of an activated allele of RAS2, both profiles of which depend on a functional PKA (Zaman et al. 2009). In addition, deletion of GPA2 is synthetically lethal with deletion of RAS2 and that lethality is suppressed by deletion of PDE2 (Kubler et al. 1997; Xue et al. 1998). All of these results are consistent with Gpa2 functioning as an activator of adenylyl cyclase.
Gpa2 physically interacts in vivo with Gpr1, a protein homologous to seven transmembrane G-protein–coupled receptors (Xue et al. 1998; Kraakman et al. 1999). Deletion of GPR1 is synthetically lethal with deletion of RAS2 and that lethality is suppressed by deletion of PDE2. These observations prompted a facile model in which Gpr1 serves as a receptor for glucose, which upon binding the ligand, stimulates activation of Gpa2, which in turn stimulates adenylyl cyclase. However, a number of observations are inconsistent with this model. First, no robust pharmacological assays defining ligands for Gpr1 have been reported. The single rather indirect assay for ligand interaction with Gpr1 suggests that Gpr1 has a weak affinity for glucose and binds with much higher affinity to nonpreferred carbon sources such as sucrose (Lemaire et al. 2004). Second, deletion of Gpr1 or Gpa2 has no effect on the transcriptional response of cells to glucose addition (Zaman et al. 2009). Finally, the PKA-dependent inactivation of fructose bisphosphatase that occurs immediately upon glucose addition to cells is retained in gpr1Δ strains (Belinchon and Gancedo 2007). Thus, Gpr1 does not appear to serve as a primary mediator in the acute response of cells to glucose addition.
Several observations have highlighted a potential connection between Gpr1/Gpa2 and Sch9, the yeast homolog of S6 kinase (see below). First, an activated allele of GPA2 elicits heat-shock sensitivity in wild-type and ras1Δ ras2Δ strains, which is suppressed in an sch9Δ background (Xue et al. 1998). This suggests that Gpa2 acts upstream of Sch9. In contrast, sch9 hypomorphic alleles increase the ability of Gpa2 to induce PKA activity, suggesting that Sch9 suppresses Gpa2 activity (Zaman et al. 2009). Moreover, gpr1Δ exhibits synthetic lethality with sch9Δ, indicating a common function for the two genes (Kraakman et al. 1999). Finally, gpr1Δ strains and strains with a hypomorphic sch9 allele exhibit an identical pattern of gene activation during growth on glycerol (Zaman et al. 2009). Thus, Gpr1 and Sch9 suppress expression of the same set of genes during growth on a nonfermentable carbon source, reinforcing the notion that these two proteins share a common function. While these observations do not coalesce into a coherent model of interaction, they do define a more direct relationship between Gpr1/Gpa2 and Sch9 than has been acknowledged.
While Gpa2 resembles the α subunit of a heterotrimeric G protein, no canonical β or γ subunits have been convincingly demonstrated to partner with Gpa2. Zeller et al. (2007) proposed that Asc1, a protein with a classical WD40 structure observed in all β subunits of heterotrimeric G proteins, serves in this capacity. Asc1 binds in vitro to Gpa2 bound to GDP but not to Gpa2 bound to GTP and deletion of ASC1 results in higher glucose stimulated adenylyl cyclase activity in vivo. However, Asc1 is a cytoplasmic protein present in vast excess of Gpa2 and participates in myriad protein complexes with a diverse set of functions in vivo, none of which are associated with Gpa2 activity. Thus, Acs1 does not fit the classic definition of a Gβ subunit and its role in Gpa2 function is still unresolved.
Gpb1/Krh2 and Gpb2/Krh1, two related proteins containing seven kelch repeats that fold into β-propeller structures like those formed by WD40 repeats, have also been proposed as β subunits partnering with Gpa2, based initially on two-hybrid interaction. However, substantial evidence has accumulated discounting Gpb1/Gpb2 as β subunits (Peeters et al. 2007), including the fact that the site on Gpa2 at which the proteins bind does not correspond to the classic Gβ-binding domain (Niranjan et al. 2007). Nonetheless, Gpb1 and Gpb2 play redundant roles in negatively regulating the activity of the Ras/PKA pathway, either by interference with the Gpr1/Gpa2 interaction (Harashima and Heitman 2005), or through stabilization of the Ras–GAP proteins, Ira1 and Ira2 (Harashima et al. 2006), or by stabilization of the interaction between the regulatory subunit, Bcy1, and the catalytic subunits, Tpk1–3, of protein kinase A (Lu and Hirsch 2005; Peeters et al. 2006; Budhwar et al. 2010), or by some combination of all three mechanisms.
One should note that the studies on Gpr1, Gpa2, and Gpb1/2 have not examined the dynamic nature of these components in the context of signal transduction. Rather, these studies exclusively provide a static view of the role of these proteins in signal output. Thus, we do not know whether these components serve a dynamic function in the signaling cascade or simply function as structural elements of the signaling machinery, imparting stability to the Ras–GAP proteins or the Bcy1–Tpk interaction, for example.
Sch9, a protein kinase B homolog:
SCH9 encodes an AGC family protein kinase homologous to the mammalian S6 kinase and the prosurvival protein kinase, Akt. It was identified as a high-copy suppressor of strains lacking protein kinase A. Overexpression of Sch9 results in essentially identical transcriptional reprogramming as does activation of protein kinase A, which suggests that the ability of Sch9 to suppress loss of PKA activity is a consequence of overlapping substrate specificities of the two kinases. In fact, their recognition motifs are quite similar and many of the identified substrates of Sch9 are substrates of PKA, although the set of substrates and the precise phosphorylation sites are not completely congruent (Huber et al. 2009; Mok et al. 2010). Nonetheless, Sch9 and PKA appear to perform similar functions in the cell by targeting overlapping substrates.
As noted below, Sch9 is activated by direct phosphorylation by TORC1 and, as such, is responsible for many of the changes in cellular protein phosphorylation elicited by TORC1 (Urban et al. 2007). Glucose also regulates Sch9 activity, both by increasing its level in the cell and by inducing its phosphorylation (Jorgensen et al. 2004). The Pkh1,2 kinases are activated by sphingolipids and phosphorylate Sch9 on its activation loop (Jacinto and Lorberg 2008). In addition, the AMP-activated protein kinase SNF1 phosphorylates Sch9 and apparently enhances its activity (Lu et al. 2011). Whether these or other kinases or phosphatases mediate glucose activation of Sch9 is not clear.
An acute increase in Sch9 activity substantially recapitulates transcriptional responses to glucose addition to cells, suggesting that Sch9 activation is sufficient to elicit the glucose transcriptional response (Zaman et al. 2009). However, inactivation of Sch9 concurrent with glucose addition does not diminish glucose-induced transcriptional changes, whereas inactivation of PKA concurrent with glucose addition reduces the magnitude of the transcriptional response by 75%. This indicates that Sch9 per se is not necessary for the glucose response, whereas PKA plays a requisite role. Nonetheless, the residual transcriptional response to glucose in the absence of PKA activity depends on Sch9 (Zaman et al. 2009). In short, while Sch9 certainly participates in nutrient signaling downstream of TORC1, it also plays a significant role in glucose regulation of cell growth.
Yak1, a proquiescence kinase:
Yak1 is a member of the conserved dual-specificity tyrosine-phosphorylation–regulated protein kinase. It functions in a PKA pathway but it inhibits rather than stimulates cell proliferation. YAK1 was identified as a loss-of-function suppressor of PKA deficiency and YAK1 overexpression inhibits cell proliferation, suggesting that it functions downstream of PKA (Garrett et al. 1991). Yak1 localizes to the nucleus following glucose depletion or rapamycin treatment but becomes phosphorylated and localized to the cytoplasm following glucose addition to cells (Moriya et al. 2001; Martin et al. 2004). Cytoplasmic 14-3-3 proteins bind phosphorylated Yak1 and inhibit its protein kinase activity. Thus, while PKA phosphorylation does not alter Yak1 kinase activity in vitro, 14-3-3 interaction resulting from PKA phosphorylation in vivo reduces its activity (Budovskaya et al. 2005; Ptacek et al. 2005; Lee et al. 2011). Consistently, Yak1 without its PKA phosphorylation sites accumulates in the nucleus, even in cells grown on glucose (Lee et al. 2011). Thus, Yak1 activity is regulated in response to glucose at least in part through PKA-dependent subcellular localization (Moriya et al. 2001).
A major downstream target of Yak1 is Pop2/Caf1, a member of the Ccr4–Caf1–Not1 deadenylation complex that controls the stability and/or translation of a variety of transcripts involved in stress response and use of alternative carbon sources (Moriya et al. 2001). Blocking Yak1 phosphorylation of Pop2 prevents cells from arresting in G1 at the end of postdiauxie prior to entry into stationary phase. Yak1 also exhibits genetic interaction with Msi1/Cac3, a high-copy suppressor of hyperactive PKA signaling and a member of the CAK chromatin deposition complex (Pratt et al. 2007). Msi1 and Yak1 work in parallel to promote cessation of growth that counteracts the effects of PKA. Yak1 impinges on the stress response pathways by directly phosphorylating the heat-shock transcription factor, Hsf1, and the major stress response transcription factors, Msn2 and Msn4 (Lee et al. 2008). Phosphorylation of Hsf1 by Yak1 increases its DNA-binding activity and Yak1 is required for full transcriptional activity of Hsf1. While Yak1 is required for full activity of Msn2/4, the absence of Yak1-induced phosphorylation does not affect nuclear localization of Msn2 in vivo or its DNA binding affinity in vitro. Finally, transcriptional profiling and genetic studies suggest that Yak1 inhibits the filamentation-antagonizing transcription factor, Sok2 (Malcher et al. 2011).
In sum, Yak1 appears to function in concert with PKA but in the opposite direction: PKA promotes cell growth and inhibits the stress response while Yak1 inhibits cell growth and stimulates the stress response. Yak1 may accomplish this by impinging directly on the PKA pathway through phosphorylation of Bcy1 (Griffioen et al. 2001) but more likely through an independent process of activating stress-responsive transcription factors and stabilizing or promoting the translation of growth-inhibitory, stationary-phase–promoting mRNAs. Moreover, glucose influences Yak1 function through a mechanism at least partially dependent on PKA. Thus, Yak1 represents a branch of the PKA pathway by which glucose regulates the growth and development commitment of the cell.
SNF1 and the use of alternative carbon sources:
The preferential use of glucose as carbon and energy source by yeast results from glucose-induced transcriptional repression of genes required for catabolism of other sugars as well as those involved in central carbon metabolism. In addition, glucose causes repression of mitochrondrial function, which is required for oxidative phosphorylation necessary for metabolism of nonfermentable carbon sources. These processes are regulated by glucose through the combined actions of the Snf1 kinase and the Hap regulatory complex.
Components of SNF1:
SNF1 was identified as a gene required for glucose repression, for growth on sucrose as sole carbon source, and for induction of invertase in response to glucose depletion. Snf1 is the catalytic subunit and founding member of the eukaryotic family of AMP-activated protein kinases (AMPKs). In mammalian cells, AMPK responds to declining energy charge of the cell by stimulating increased glucose uptake and oxidation, increased fatty acid oxidation, inhibition of anabolic reactions, and stimulation of reactions that generate ATP. Thus, AMPK serves as a guardian of energy homeostasis in cells, promoting increased energy production and reduced energy demand by a multiplicity of means when energy reserves are depleted (Hardie et al. 1998, 2011). In yeast, Snf1 kinase performs similar functions but may do so in direct response to declining glucose levels rather than energy charge, reflecting the fact that yeast cells assess their nutrient sufficiency predominantly through their perception of glucose rather than their metabolism of it.
Like other members of the AMPK family, SNF1 protein kinase is a heterotrimer comprising the Snf1 catalytic (α) subunit, a regulatory (γ) subunit, Snf4, and one of three β subunits—Gal83, Sip1 or Sip2—that function as scaffold and localization determinants. In this review, I will refer to the complex as SNF1, distinct from the catalytic subunit Snf1 and the gene SNF1. The Snf1 catalytic subunit contains an N-terminal kinase domain and a C-terminal autoinhibitory domain. In mammalian AMPKs, binding of AMP to the γ subunit stimulates kinase activity via allosteric alteration of interaction of the autoinhibitory domain with the kinase domain (Chen et al. 2009). Snf4 is required for SNF function in yeast cells but deletion of the autoinhibitory domain of Snf1 eliminates the requirement for Snf4 for kinase activity in vivo and in vitro, suggesting that the primary function of Snf4 is to alleviate Snf1 autoinhibition.
Snf4 consists of two pairs of repeats, termed Bateman domains, which in other proteins bind adenosine derivatives. The structure of this domain in Snf4 is quite similar to the Schizosaccharomyces pombe γ subunit, which binds a single molecule of AMP or ATP (Townley and Shapiro 2007). Recent results have shown that amino acid substitutions within the Bateman domain of Snf4, analogous to some disease causing activating alleles in the human AMPK γ subunit, alleviate to some extent the inhibitory effects of glucose on SNF1 activity (Momcilovic et al. 2008). This would suggest that allosteric changes in Snf4 resulting from these substitutions can result in reduced deactivation of the catalytic subunit by glucose. However, AMP fails to activate SNF1 in vitro, suggesting that AMP does not bind or stimulate SNF1 in vivo (Mitchelhill et al. 1994; Woods et al. 1994; Wilson et al. 1996). Rather, ADP binds to Snf4 and, at least in vitro, protects against dephosphorylation of Thr210 (see below) (Mayer et al. 2011). But, since 2-deoxyglucose, which can be phosphorylated by hexokinase but cannot be further metabolized, inhibits SNF1 activity in vivo, glucose does not have to be extensively metabolized to affect SNF1 function. Finally, glucose regulates phosphorylation of the Snf1 activation domain in vivo (see below) even in the absence of Snf4 and the Snf1 autoinhibitory domain (Jiang and Carlson 1996; Leech et al. 2003). Thus, while Snf4 is required to alleviate autoinhibition of Snf1 and mutations in Snf4 can attenuate glucose inhibition of SNF1, Snf4 does not seem to appreciably regulate SNF1 in response to energy charge in the cell.
Activation of SNF1 kinase activity results from phosphorylation of threonine 210 in the activation loop of Snf1. Three kinases—Elm1, Tos3, and Sak1—serve as redundant SNF1-activating kinases (Hong et al. 2003; Nath et al. 2003; Sutherland et al. 2003). In mammalian cells, this function is performed redundantly by LKB1, Ca2+/calmodulin-dependent kinase (CaCDK) and TGFβ-activated kinase, the identities of which were revealed by heterologous complementation in yeast (Hong et al. 2005; Momcilovic et al. 2006). The action of the three SNF1-activating kinases is counteracted by the essential protein phosphatase 1, Glc7, in conjunction with its specificity subunit, Reg1 (Tu and Carlson 1995). Low glucose levels correlate with increased phosphorylation of Thr210 and enhanced SNF1 activity (McCartney and Schmidt 2001). Several results suggest that glucose does not act through the upstream kinases: SNF1 activity exhibits normal regulation in strains in which the three yeast kinases are functionally replaced by mammalian LKB1. Moreover, the three upstream kinases exhibit the same activity in extracts of cells grown in glucose-limited or glucose-replete media (Hong et al. 2005; Rubenstein et al. 2008). Unlike mammalian AMPK, AMP does not stimulate SNF1 in yeast, although ADP binding to Snf4 protects Thr210 from dephosphorylation, at least in vitro (Mayer et al. 2011). Rather, glucose must regulate SNF1 activity either by inhibiting one or more of the upstream kinases, or by activating the Reg1/Glc7 phosphatase, or by rendering Thr210 more accessible to dephosphorylation. Finally, while high glucose levels accelerate the rate of Thr210 dephosphorylation in vivo, Reg1/Glc7 activity in vivo appears unaffected by changes in glucose levels (Rubenstein et al. 2008). Thus, glucose may regulate SNF1 activity by modifying the accessibility of the complex to the Reg1/Glc7 phosphatase, perhaps through reduction in ADP levels or through modulation of the interaction between SNF1 and Reg1 (Dombek et al. 2004; Rubenstein et al. 2008; von Plehwe et al. 2009; Mayer et al. 2011).
While Snf1 activation has been studied predominantly in the context of glucose repression, Snf1 is phosphorylated and activated in response to a number of environmental stresses. Alkaline pH, high sodium chloride, or oxidative agents, but not high sorbitol or heat shock, result in increased Thr210 phosphorylation and SNF1 activity as well as nuclear relocalization (Hong and Carlson 2007). All three upstream kinases contribute to this stress-induced phosphorylation with Sak1 playing the predominant role. However, as with glucose regulation of SNF1 activity, SNF1 responds to these stresses even in elm1Δ sak1Δ tos3Δ strains expressing mammalian CaCDK. Thus, activation of SNF1 in response to stress appears to result from inactivation of Reg1/Glc7 phosphatase rather than activation of the upstream kinase. Finally, SNF1 is activated by Thr210 phosphorylation in response to nitrogen starvation and TORC1 inactivation (see below) (Orlova et al. 2006). In this case, phosphorylation is solely dependent on Sak1, suggesting that TORC1 might regulate this Snf1-activating kinase directly (Orlova et al. 2010).
The β subunits all contain domains for binding Snf1 and Snf4 and as such provide a scaffold for assembly of the kinase complex. In addition, both Gal83 and Sip2 contain a glycogen binding domain, although Gal83 binds glycogen avidly, while Sip2 does so only weakly. Mutations within the glycogen-binding domain of Gal83 or deletion of the domain alleviate glucose-induced inhibition of SNF1 kinase activity in vivo although elimination of glycogen in the cell does not (Momcilovic et al. 2008). This suggests that this domain may alter the structure of the complex in a way that allows glucose-induced inhibition of kinase activity but does not provide a means for regulation of the complex in response to glycogen levels.
The β subunits confer distinct functions and subcelllular localizations of the SNF1 complex (Schmidt and McCartney 2000; Vincent et al. 2001). In glucose grown cells, all three complexes reside in the cytoplasm. In limiting glucose, Gal83-containing SNF1 complexes relocate to the nucleus, where they participate in transcriptional activation; Sip1-containing complexes relocate to the vacuolar periphery; and the Sip2-containing complexes remain in the cytoplasm. In response to alkaline stress, SNF1 relocates to the nucleus, while in response to salt stress, it remains in the cytoplasm. This suggests that regulation of subcellular location may contribute to the specificity of SNF1 action.
While β subunits usually promote increased SNF1 activity toward selected substrates, Sip2 appears to function as an inhibitor of SNF1 function, at least in older yeast cells (Ashrafi et al. 2000). SNF1 activity increases in older cells, resulting in diminished replicative aging; sip2 mutants exhibit shortened replicative lifespan, an effect that is reversed by concurrently deleting SNF1, suggesting that Sip2 inhibits Snf1 function in older cells. Recent results demonstrate that Sip2 is acetylated in vivo by the NuA4 acetyl transferase complex, a modification that enhances its interaction with Snf1 and increases replicative lifespan, likely through inhibition of SNF1 activity (Lu et al. 2011). SNF1 phosphorylates and activates Sch9, which serves as the critical downstream target in SNF1’s effect on replicative aging in older cells, and acetylated Sip2 diminishes the activity of SNF1 toward Sch9. Thus, Sip2 inhibits SNF1, reducing activation of Sch9 and extending replicative lifespan.
Transcriptional regulation by SNF1:
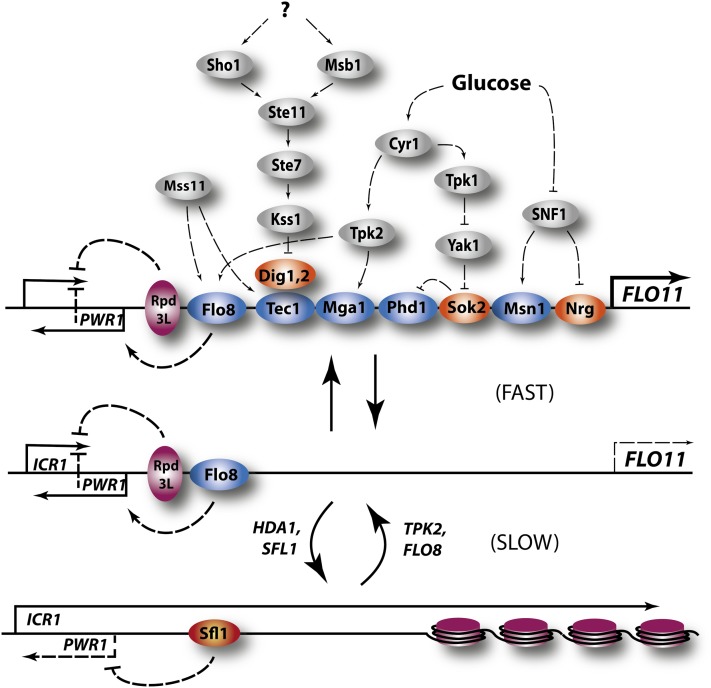
Activated SNF1 promotes expression of hundreds of genes involved in use of alternate carbon sources through a variety of transcription factors and promotes repression of a number of genes involved in amino acid metabolism through Gcn4 (Figure 3) (Young et al. 2003; Shirra et al. 2008; Zaman et al. 2009). Genes required for metabolism of alternative sugars, such as sucrose, galactose, and maltose, respond to Snf1 through the Mig1 transcriptional repressor, a C2H2 zinc finger protein that binds to a GC-rich consensus sequence (reviewed in Schuller 2003). In cells grown in the absence of glucose, Snf1 phosphorylates Mig1 to inhibit Mig1’s repressor activity. In the presence of glucose, Mig1 becomes dephosphorylated and localizes to the nucleus repressing expression of target genes such as SUC2. Mig1 acts as a repressor in association with Hxk2, one of the two yeast hexokinases (Ahuatzi et al. 2004, 2007). Hxk2 forms a complex in vitro with a SUC2 DNA and Mig1, suggesting that Hxk2 interacts with Mig1 as part of the repressor complex on the SUC2 promoter. Moreover, Hxk2 interacts specifically through the S311 residue of Mig1, mutation to a nonphosphorylatable form of which results in constitutive localization of Mig1 to the nucleus and constitutive inhibition of SUC2 expression. Finally, certain mutants of Hxk2 defective in catalytic activity retain full corepressor activity and certain mutants defective in repressor activity retain catalytic activity (Pelaez et al. 2010). Thus, Hxk2 participates in regulation independently of its metabolic activity.
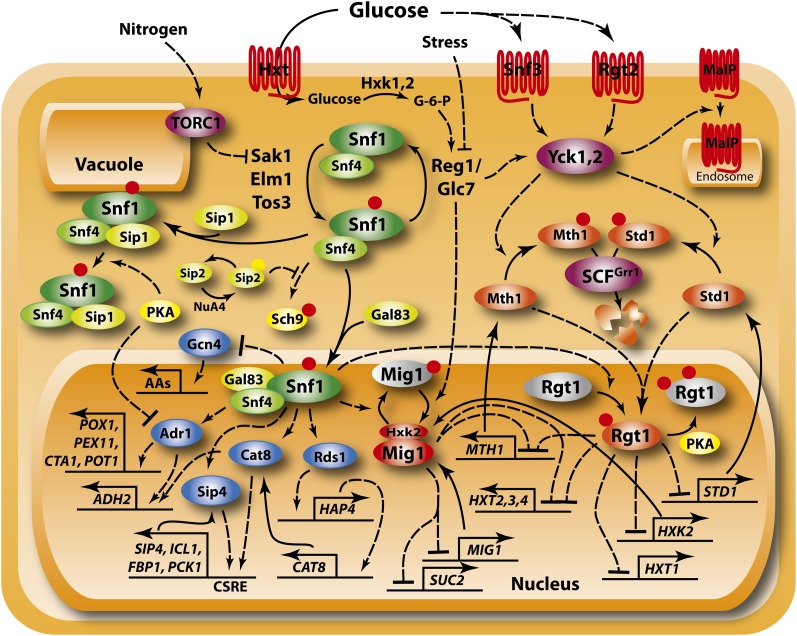
Figure 3 .
The Snf1 and Rgt pathways. The Snf1 (green and yellow icons) and Rgt (purple and orange icons) are interlocking pathways that regulate use of alternate carbon sources, primarily through regulation of a constellation of transcriptional activators (blue icons) and repressors (red and orange icons). Rgt1 responds to glucose levels through two membrane glucose sensors. Snf1 responds primarily to glucose through modulation of the Reg1/Glc7 protein phosphatase 1 (PP1), although stress and nitrogen levels also impinge on Snf1 activity through distinct routes. Snf1 also represses amino acid biosynthetic genes (AAs) through inhibition of Gcn4 translation. The Snf1/Snf4 holoenzyme acquires substrate specificity through interaction with one of three distinct β subunits, Gal83, Sip1, or Sip2. Acetylation (yellow dot) of Sip2, catalyzed by NuA4 and reversed by Rpd3, stimulates associated between Sip2 and Snf1, an interaction that blocks activation of Sch9. Finally, PP1 stimulates internalization of the maltose permease (MalP) in response to glucose through the action of yeast casein kinases 1 and 2 (Yck1,2).
Snf1 regulates expression of genes involved in ethanol metabolism and β oxidation of fatty acids through modulation of the Adr1 transcription factor (Ratnakumar and Young 2010). Deletion of ADR1 reduced the expression of ∼100 genes in cells grown on low glucose (Young et al. 2003). This study showed that Adr1 also affected expression of genes in other functions, such as amino acid transport and metabolism, meiosis, and sporulation. However, since only 30 genes are tightly bound by Adr1 in cells grown in glucose-free media, altered regulation of most genes in an adr1 could be the consequence of secondary regulatory or metabolic effects (Tachibana et al. 2005; Zaman et al. 2009).
Adr1 is negatively regulated by phosphorylation on serine 230 in glucose-grown cells and activated by dephosphorylation of that site in a Snf1-dependent manner in cells grown in the absence of glucose (reviewed in Schuller 2003). While PKA and CaCDK can phosphorylate this site in vitro, neither is essential for its phosphorylation in vivo, suggesting that redundant and/or some other kinases serve in that capacity (Ratnakumar et al. 2009). Moreover, the mechanism by which Snf1 induces dephosphorylation of S230 is unknown. Adr1 is also under negative regulation of Reg1, as deletion of REG1 increases the protein level of Adr1 and leads to induction of Adr1-regulated genes, such as ADH2 (Dombek et al. 2004). The yeast 14-3-3 proteins, Bmh1 and Bmh2, likely act in a pathway parallel to Reg1 to inhibit expression of Adr1-regulated genes. Bmh1 and Bmh2 bind to Adr1 phosphorylated on S230 (Parua et al. 2010) and expression of ADH2 under repressed conditions is increased in a bmh1bmh2 strain and even further increased in a reg1bmh1bmh2 strain. Thus, Adr1 is sensitive to a number of glucose-dependent inputs.
Several unrelated transcription factors, including Cat8, Sip4, and Rds2, activate expression of genes required for gluconeogenesis during growth in the absence of glucose by binding carbon source response elements (CSRE). Derepression of genes having CSRE motifs is completely abolished in cat8sip4 mutants, suggesting that these two proteins are the major activators (reviewed in Schuller 2003; Turcotte et al. 2010). However, Cat8 and Sip4 do not equally contribute to activation of genes in the absence of glucose: cat8 cells cannot grow on nonfermentable carbon sources, whereas sip4 mutants can. This hierarchy is further supported by the fact that Sip4 has much stricter requirement for the consensus CSRE motifs than does Cat8 (Roth et al. 2004). Of the 255 genes whose expression is reduced in the cat8 relative to CAT8 in low glucose media, only 48 are bound by Cat8 in vivo, again suggesting a large contribution of secondary events in microarray studies. During growth of cells in ethanol, Rds2 binds to a set of CSRE-containing genes distinct from, but partially overlapping with, those bound by Cat8. Rds2 activity as a transcriptional activator is enhanced during growth on nonfermentable carbon sources and correlates with Snf1-dependent hyperphosphorylation. Similarly, Sip4 responds to glucose starvation through Gal83-mediated phosphorylation by Snf1 (Vincent and Carlson 1999). CAT8 transcription is inhibited by Mig1 and activated by Hap2/3/4/5, while Rds2 activates expression of Hap4. Thus, the induction of gluconeogenic, TCA cycle, and glyoxylate shunt genes in response to glucose limitation involves a complex interplay of interacting transcription factors downstream of SNF1.
SNF1 protein kinase complex regulates certain stress response genes during carbon source downshift. Phosphorylation of Hsf1 and its subsequent binding to heat-shock elements (HSE) and activation of genes in response to carbon stress, such as HSP82, CUP1, HSP30, and SSA3, depend in part on SNF1 (Sanz 2003; Hahn and Thiele 2004). SNF1 also attenuates the Msn2 response to carbon stress. Msn2 is dephosphorylated by Reg1–Glc7 immediately following glucose depletion and localizes to the nucleus to induce expression of target genes such as CTT1 (De Wever et al. 2005). However, long-term carbon stress induces rephosphorylation of Msn2 in a SNF1-dependent manner leading to relocalization of Msn2 to the cytoplasm and inhibition of CTT1 expression (De Wever et al. 2005). This suggests that SNF1 is involved in long-term adaptation to carbon stress by negatively regulating Msn2 transcriptional activity.
SNF1 also affects gene expression by stimulating chromatin remodeling. Glucose depletion yields Snf1-dependent phosphorylation of S10 on histone H3 at the INO1 promoter (Lo et al. 2001, 2005), resulting in recruitment of the SAGA complex and acetylation of histone H3 K14. Glucose depletion results in a similar Snf1-dependent recruitment of the SAGA complex to the HXT2 and HXT4 promoters under glucose limitation (van Oevelen et al. 2006). Moreover, SNF1 phosphorylates Gcn5 in vitro, the histone acetyl transferase component of SAGA, and stimulates its activity (Liu et al. 2010). Thus, Snf1 promotes transcriptional activation through both mobilization of transcription factors and remodeling of chromatin structure of target promoters.
Finally, SNF1 impinges on the Gcn4 control of amino acid biosynthesis genes (Ljungdahl and Daignan-Fornier, 2012). In addition to repression of the genes involved in carbon metabolism noted above, inactivation of Snf1 unexpectedly results in induction of dozens of genes involved in amino acid metabolism regulated by Gcn4 (Shirra et al. 2008; Zaman et al. 2009). This suggests that under glucose-depleted conditions, SNF1 inhibits Gcn4 production or transcriptional activation. Subsequent studies have indicated that SNF1 plays additional roles in activating Gcn4, depending on the condition: under amino-acid-limiting conditions in the presence of glucose, SNF1 collaborates with uncharged tRNA to activate Gcn2, which ultimately leads to increased Gcn4 translation through increased phosphorylation of eIF2α. In glucose-limiting conditions, active SNF1 inhibits two protein phosphatases responsible for dephosphorylating eIF2α, Sit4 (see below), and Glc7. This SNF1-promoted increase in eIF2α phosphorylation also results in increased Gcn4 translation (Cherkasova et al. 2010). Thus, SNF1 appears to both stimulate and inhibit Gcn4, perhaps indicating a subtle interplay between energy homeostasis and amino acid biosynthesis coordinated by Snf1.
Metabolic regulation by SNF1:
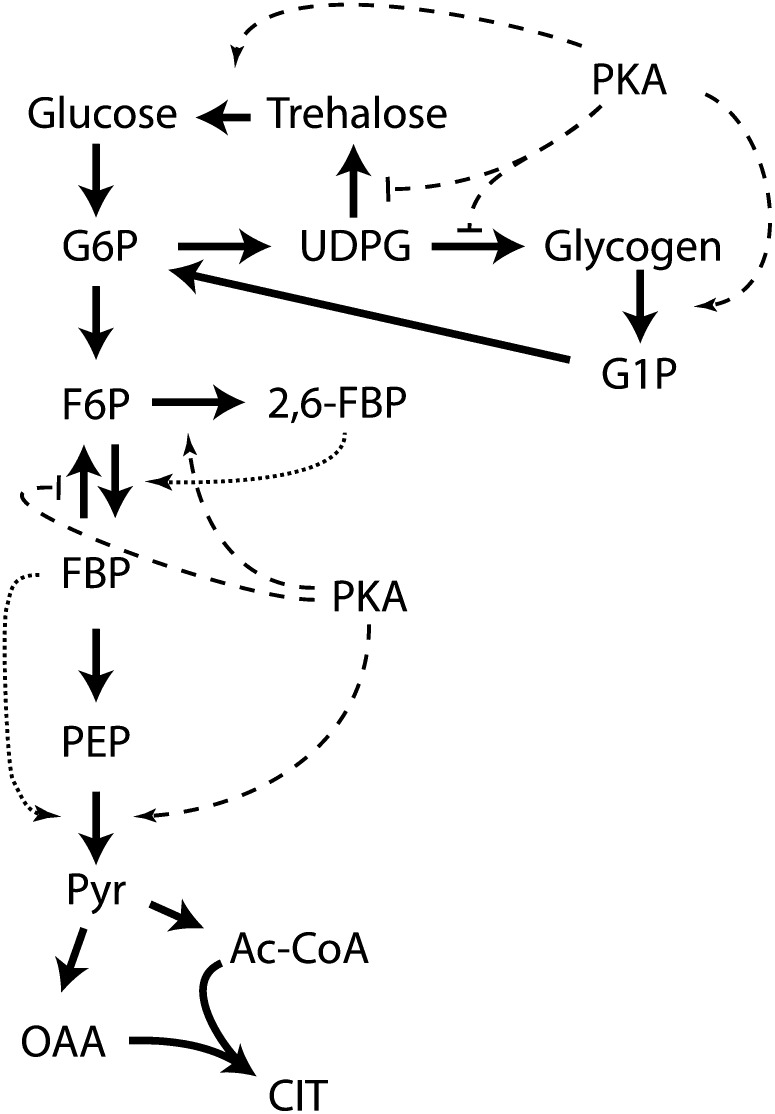
While most of the studies of SNF1 have focused on its transcriptional targets, SNF1 also modulates energy consumption and generation through direct regulation of metabolic activity, most notably of lipid biosynthesis and catabolism. SNF1 directly phosphorylates and inactivates acetyl coenzyme A (acetyl-CoA) carboxylase (Acc1), the enzyme that catalyzes the rate-limiting step in fatty acid biosynthesis, and thus minimize lipid biosynthesis in carbon-limiting conditions (Woods et al. 1994). SNF1 promotes fatty acid degradation through β oxidation in part by promoting biogenesis of peroxisomes (Hiltunen et al. 2003; Ratnakumar and Young 2010). Whether SNF1 has an additional direct role in modulating the biochemical activity of the peroxisome is not known, but free fatty acids accumulate in snf1 strains under glucose-limiting conditions, demonstrating the requirement for SNF1 in stimulating β oxidation to generate energy under nutrient-limited conditions (Usaite et al. 2009).
In sum, SNF1 couples the absence of glucose or other stresses to the suppression of energy-consuming activities and the induction of energy-generating processes. This is accomplished primarily through induction of a limited number of genes required for metabolism of carbon sources other than glucose as well as activation of genes required for gluconeogenesis and fatty acid oxidation. In the absence of SNF1 function, ∼400 genes normally induced by glucose depletion show diminished induction, although only 10% of these are direct targets of transcription factors regulated by SNF1. In addition, SNF1 likely affects the metabolic flux in the cell through modulation of the activities of key biosynthetic and catabolic enzymes, particularly in fatty acid metabolism. Unlike mammalian cells, yeast cells regulate Snf1 activity not in response to energy charge but rather through phosphorylation of the activation loop catalyzed redundantly by several upstream kinases and counteracted by protein phosphatase 1, albeit recent work has implicated ADP as a potential modulator of SNF1 activity. Current evidence supports the conclusion that glucose impinges on SNF1 through modulation of the phosphatase. We still do not understand how glucose alters the activity of the phosphatase, although glucose has to be phosphorylated, albeit not metabolized, to affect SNF1 function.
The HAP2/3/4/5 complex and mitochrondrial biogenesis:
A number of genes, particularly those involved in respiration and oxidative phosphorylation, are repressed by glucose independently of PKA and Snf1. Many of these are regulated by the Hap2/3/4/5 transcription complex, suggesting that the Hap complex may provide an independent route for glucose regulation of gene expression (Zaman et al. 2009). The Hap2/3/4/5 complex plays a central role in converting cells from fermentative to respiratory growth following the diauxic shift by inducing genes required for mitochondrial function upon glucose depletion. Hap2, -3, and -5 form a DNA-binding complex and are constitutively expressed. Hap4 provides the activation domain of the complex and its levels increase upon glucose depletion (Forsburg and Guarente 1989; Derisi et al. 1997). Increased expression of Hap4 alone yields induction of those genes under control of the complex (Lascaris et al. 2003). While Hap4 could be a target of Rds2 transcriptional induction in response to SNF1 activation, the fact that induction of Hap complex responsive genes is independent of SNF1 activity suggests an independent mechanism for Hap complex activation. The nature of the connection between glucose depletion and Hap complex activation remains to be determined.
The Rgt network and glucose transport:
The expression of many hexose transporter genes (HXTs) is precisely tuned to glucose levels available to cells to insure that the glucose transporters produced provide the most efficient import of available glucose, over a wide range of external glucose concentrations (Kaniak et al. 2004; Zaman et al. 2009). This tuning is achieved through two intertwined signaling networks, one mediated by Snf1 and one mediated by Rgt1 (Figure 3). Rgt1 is a zinc cluster DNA-binding protein that, in association with corepressors, Mth1 and Std1, represses HXT gene expression, such as HXT1–4, as well as the hexokinase gene, HXK2 (Lakshmanan et al. 2003; Mosley et al. 2003). The corepressors, Mth1 and Std1, play partially redundant roles in regulation: they each bind to a common site on Rgt1 to suppress transcriptional activation and block access to PKA, whose hyperphosphorylation of Rgt1 elicits its eviction from promoters (Palomino et al. 2006). Rgt repression activity is alleviated by binding of external glucose to two membrane-spanning glucose sensors, Snf3 and Rgt2. These sensors likely detect the relative external-to-internal glucose concentrations (Wu et al. 2006; Karhumaa et al. 2010). Glucose activation of the sensors induces functional recruitment of Mth1 and Std1 to the plasma membrane, where they are phosphorylated by casein kinases, Yck1 and Yck2. Once phosphorylated, the corepressors are targeted by the SCFGrr1 E2/E3 ubiquitin-conjugating complex for degradation by the proteosome (Schmidt et al. 1999; Flick et al. 2003; Moriya and Johnston 2004; Spielewoy et al. 2004). Elimination of these corepressors by proteolysis exposes Rgt1 to phosphorylation and alleviates its repressive activity (Palomino et al. 2006).
The repression activity of Rgt1 is stimulated by direct phosphorylation by Snf1. In contrast, some of the hexose transporter genes are repressed by Mig1, whose nuclear localization is blocked by SNF1 phosphorylation. Thus, SNF1 both promotes and attenuates repression. Moreover, STD1 expression is autoregulated by the Rgt1 network, and thus induced by high glucose, whereas MTH1 expression is repressed at high glucose by the Snf1-regulated Mig1 repressor. These observations prompt a model in which Mth1 serves primarily to maintain repression, while Std1 functions predominantly in establishment of repression during transition to the absence of glucose (Kim et al. 2006; Sabina and Johnston 2009). This complex interplay between the components of the Rgt network and Snf1/Mig1 provides a graded derepression of the different hexose transporters in response to different glucose levels, such that cells express only those transporters with the appropriate affinity for the available glucose (Johnston and Kim 2005). Albeit quite complex, with both feed-forward and feed-back regulatory loops, this network is sufficiently well defined to allow predictive modeling of its behavior both in a steady state and kinetic representations (Figure 3) (Kuttykrishnan et al. 2010).
Protein phosphatase 1:
While assignment of direct roles of protein phosphatases in various biological processes has been notoriously difficult, growing evidence suggests that the Glc7 protein phosphatase 1 plays a central role in glucose signaling. Glc7, which encodes the sole and essential protein phosphatase 1 in yeast, has little specificity on its own but associates with a large number of regulatory subunits that target its activity to different subsets of proteins. One such regulatory subunit, Reg1, binds to Glc7 to promote glucose repression predominantly through inactivation of Snf1 by dephosphorylation of its activation loop leading to activation of Mig1 (Tu and Carlson 1995). Consistent with this model, deletion of REG1 results in constitutive activation of Snf1 and hyperphosphorylation of its activation loop (McCartney and Schmidt 2001). As noted above, glucose stimulates dephosphorylation of Snf1 either by direct activation of Reg1/Glc7 or by promoting the productive interaction of Snf1 with Reg1/Glc7.
Glucose induces internalization and degradation of maltose permeases through a process that requires Yck1,2-induced phosphorylation of the permeases. Surprisingly, phosphorylation and degradation of the permeases also require Reg1/Glc7 acting upstream of the Yck1,2 kinases: reg1 mutants are defective in glucose-induced internalization and degradation of maltose permeases, a defect that is suppressed by overexpression of Yck1 (Gadura et al. 2006). These results are consistent with the idea that Reg1/Glc7 enhances Yck1,2 activity, although a mechanistic link is currently lacking. Rgt2 is also required for glucose-induced maltose permease turnover: rgt2 mutants exhibit reduced internalization and an RGT2 constitutive allele induces turnover even in the absence of glucose (Jiang et al. 1997; Gadura et al. 2006). These observations suggest that glucose impinges on maltose permease internalization and degradation through two routes, one by direct binding to Rgt2 and one through activation of Yck1,2 via Reg1/Glc7. Whether this second route involves direct activation of Reg1/Glc7 by glucose has not been established but is consistent with the observations.
Msn2 and Msn4, the major stress-responsive transcription factors, are regulated predominantly through their nuclear localization as a result of phosphorylation of a nuclear localization site (NLS) on the proteins (Gorner et al. 2002). Phosphorylation of this domain, catalyzed by PKA, restricts the proteins to the cytoplasm while dephosphorylation of the domain, catalyzed by Glc7, renders the site functional and promotes nuclear entry and activation of stress-responsive genes (Gorner et al. 1998, 2002). Dephosphorylation of the NLS occurs much too quickly upon glucose downshift to be explained solely as an inhibition of PKA activity (De Wever et al. 2005). Rather, the kinetics suggest that glucose depletion induces Msn2 nuclear localization through activation of Glc7. Neither deletion of Reg1 nor of Bub14, another regulatory subunit of Glc7 implicated in activation of Msn2 upon diauxic shift (Lenssen et al. 2005), alleviated the glucose-depletion–induced nuclear localization of Msn2 (De Wever et al. 2005). Accordingly, some as yet unidentified specificity subunit likely mediates the effects of glucose depletion on activation of Glc7 toward Msn2.
There are certainly other glucose-regulated processes, such as glycogen and trehalose accumulation and, as noted above, eIF2a phosphorylation, in which protein phosphatase plays a role, although whether as a direct conduit of the glucose signal or as a foil to glucose-regulated kinases remains to be determined.
Regulatory networks responsive to nitrogen source
Nitrogen regulation:
Growth control:
Yeast cells recognize the nature and availability of nitrogen compounds and actively adjust their transcriptional, metabolic, and biosynthetic capabilities to match that perception. When nitrogen is limiting, cells slow their growth, primarily through reduction in ribosomal biogenesis and translation, resulting in expansion of the G1 phase of the cell cycle (Brauer et al. 2008). In the extreme case of nitrogen depletion, cells cease growing, even with all other nutrients available in excess, and enter a nitrogen-specific quiescent state (Klosinska et al. 2011). Unlike auxotrophic cells starved for their required amino acid, such quiescent cells retain viability for an extended period of time and suppress catabolism in a way that prevents consumption of ambient glucose in the medium (Brauer et al. 2008). Thus, yeast cells couple their synthetic capacity and growth rate to the quality and amount of available metabolizable nitrogen.
Nitrogen catabolite repression:
While yeast cells can use a variety of nitrogen-containing compounds as sole nitrogen source, they exhibit a hierarchical preference for those sources. Most laboratory strains prefer glutamine or ammonia but will use other nitrogen sources, albeit with a reduced growth rate. Moreover, yeast exhibit nitrogen catabolite repression (NCR) in which preferred nitrogen sources repress expression of genes required for uptake and catabolism of less preferred nitrogen sources (Magasanik and Kaiser 2002). Nitrogen catabolite repression is further manifest by post-translational regulation of the spectrum of amino acid permeases residing in the plasma membrane, such that the high capacity general amino acid permease, Gap1, is maintained at the cell surface only under poor nitrogen conditions (Magasanik and Kaiser 2002). Finally, availability of a readily metabolizable nitrogen source suppresses the process of autophagy, by which the cell delivers cytoplasmic macromolecular components to the vacuole for proteolytic recycling of the component parts (Yang and Klionsky 2009). Thus, nitrogen accessibility regulates metabolism, growth, transcription, post-transcriptional protein sorting, and protein turnover in yeast.
The addition of glutamine or ammonia to cells growing on a poor nitrogen source results in a number of transcriptional changes, including induction of genes required for growth and repression of genes for use of poorly metabolized nitrogen sources. This latter category comprises the ∼90 genes subject to NCR, which are regulated by an interplay of four GATA family zinc-finger transcription factors: two transcriptional activators, Gln3 and Gat1 (Nil1 and Mep80), and two repressors, Dal80 and Gzf3 (Deh1 and Nil2) (Cooper 2002; Magasanik and Kaiser 2002; Scherens et al. 2006). Cells regulate NCR genes primarily by modulating subcellular localization of the transcriptional activators: during growth on poor nitrogen sources, Gln3 and Gat1 localize to the nucleus where they bind to GATA sequences in promoters of NCR genes, while during growth on ammonium or glutamine, the transcription factors reside in the cytoplasm. Ure2 serves as an anchor to sequester Gln3 in the cytoplasm: Gln3 resides in the nucleus and fully activates NCR transcription in a ure2 mutant, regardless of nitrogen source. This observation demonstrates not only that Ure2 serves as a cytoplasmic anchor for Gln3 but also that nitrogen deprivation acts on Gln3 solely to liberate it from sequestration by Ure2. Gat1 does not localize to the nucleus in a ure2 mutant. This suggests that a separate as yet unidentified protein may anchor Gat1 in the cytoplasm in cells grown on glutamine or ammonia or that Gat1 phosphorylation directly regulates its interaction with the nuclear import machinery.
Retrograde regulation:
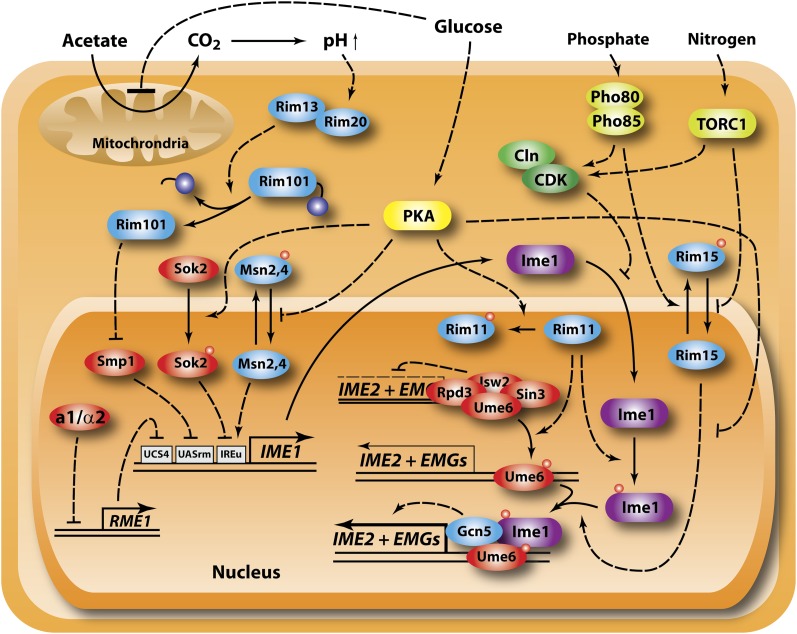
Yeast cells assimilate nitrogen from sources other than glutamate and glutamine by converting them to ammonium and then condensing the ammonia with α-ketoglutarate to form glutamate. α-ketoglutarate can be generated from pyruvate and acetyl-CoA by an anapleuotic pathway catalyzed by the first three enzymes of the citric acid cycle. However, since genes of the citric acid cycle are repressed during growth on glucose, genes encoding enzymes of this portion of the citric acid cycle are specifically upregulated during growth on certain poor nitrogen sources by activators of the RTG pathway, which responds both to mitochondrial dysfunction and to growth on nitrogen sources requiring α-ketoglutarate for assimilation (Liu and Butow 1999). In this way, the RTG pathway provides a means of ammonium assimilation from poor nitrogen sources and a source of glutamate in the absence of mitochrondrial function (Figure 4).
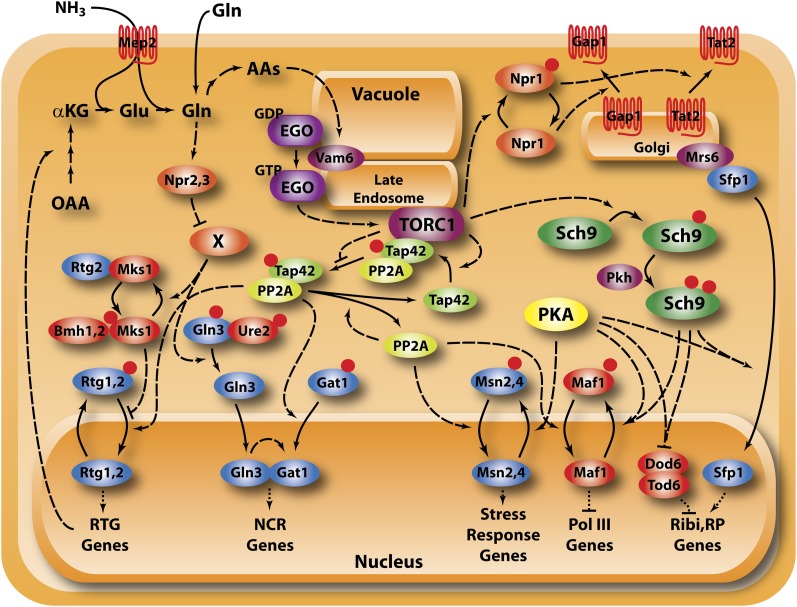
Figure 4 .
TORC1 and nitrogen regulation. Two pathways, one mediated by TORC1 and a second less well-defined nitrogen catabolite repression pathway, adjust growth as well as expression of genes required for use of alternate nitrogen sources in response to the quality and quantity of available nitrogen sources through regulation of transcriptional activators (blue icons) and repressors (red icons). TORC1 likely responds to intracellular amino acid levels sensed through the Ego complex and regulates growth primarily through Sch9, regulates stress, and alternative nitrogen source through protein phosphatase 2A and regulates permease sorting through Npr1. Npr2/3 lie upstream of NCR gene expression but whether they regulate TORC1 or the ill-defined NCR pathway is not clear.
The RTG regulatory pathway consists of four positive regulators—Rtg1, Rtg3, Rtg2, and Grr1 and four negative regulators—Mks1, Bmh1, Bmh2, and Lst8 (Liu and Butow 2006). Rtg1 and Rtg3 form a heterodimeric transcriptional activator whose nuclear localization is regulated by the other components of the pathway in response to mitochrondrial integrity and nitrogen availability. When mitochrondria are functional and sufficient nitrogen is available, the transcription factors are cytoplasmic; disruption of mitochrondrial function or nitrogen depletion results in nuclear localization of the factors and subsequent transcriptional activation of target genes. Regulation of the nuclear/cytoplasmic trafficking of Rtg1/Rtg3 involves complex interactions among Mks1, Rtg2, and Bmh1/2 (Dilova et al. 2004). When phosphorylated, Mks1 complexes with Bmh1/2 to form an anchor that sequesters Rtg3/Rtg1 in the cytoplasm. Rtg2 can compete for Bmh1/2 binding to Mks1 and thereby relieve the cytoplasmic sequestration and promote nuclear entry and transcriptional activation by Rtg1/3. Release of Mks1 from Bmh1/2 is associated with reduced phosphorylation of Mks1. Grr1, the SCF-targeting subunit, promotes ubiquitination and subsequent degradation of Mks1, providing a long-term modulation of the pathway, while Lst8, a subunit of the TOR complexes, renders the RTG pathway sensitive to Tor inhibition (Figure 4).
Nitrogen regulatory pathways:
At least two pathways mediate the response of yeast cells to nitrogen availability. The rapamycin-sensitive TORC1 complex, universally conserved among eukaryotic cells, is the central mediator and coordinator of physiological responses of the cell to changes in nitrogen source and availability (De Virgilio and Loewith 2006a). The yeast TOR complex I (TORC1) comprises a phosphatidylinsotiol kinase-related protein kinase, Tor1 (or in its absence, Tor2), Kog1 (homolog of mammalian raptor), Lst8, and Tco89 and exerts its biological function as a protein kinase. In yeast cells, TORC1 responds predominantly to nitrogen availability, likely sensed as the level of intracellular amino acids. The primary evidence positing a central role for TORC1 in nitrogen signaling is the strong correlation in the responses of cells to nitrogen starvation and the responses of cells to rapamycin addition, which specifically inhibits TORC1 activity (Cardenas et al. 1999; Bertram et al. 2000; Shamji et al. 2000). However, the fact that rapamycin addition does not fully phenocopy nitrogen depletion, particularly with regard to retrograde transcription and nitrogen catabolite repression (Tate and Cooper 2003; Tate et al. 2009, 2010), demands the existence of at least one other nitrogen signaling pathway. Neither the constituents nor the structure of that pathway has been defined.
The TORC1 pathway and cellular growth control:
Regulation of TORC1:
In mammalian cells, TORC1 provides a nexus for integrating energy charge, growth factor signaling, amino acid availability, and other nutritional inputs. Signaling pathways for energy charge and growth factors impinge on TORC1 through the heterodimeric Tsc1/2 tubular sclerosis complex, which stimulates the GTPase activity of the Rheb small G protein, whose binding to TORC1 in its GTP-bound state is necessary for TORC1 kinase activity (Sarbassov et al. 2005). However, stimulation of mammalian TORC1 by amino acids occurs independently of Tsc1/2 and is mediated instead by a heterodimer of two small GTP-binding protein, consisting of either RagA or RagB and either RagC or RagD. The Rag complex, activated by the presence of amino acids, promotes relocalization of TORC1 from discrete cytoplasmic sites to a late endosomal or lysosomal compartment at which Rheb resides (Sancak et al. 2010). Thus, amino acid availability regulates mammalian TORC1 in a manner distinct from other inputs.
The yeast TORC1 responds primarily to the quality and amount of nitrogen in the environment (Figure 4). Deceased TORC1 activity occurs upon nitrogen starvation or downshift and increased activity results from nitrogen source upshifts or from cycloheximide treatment, which causes an increase in intracellular amino acids as a result of diminished protein synthesis (Binda et al. 2009). Previous results have suggested that the quantity and quality of nitrogen source is perceived as the level of intracellular glutamine: mutations in GLN1 that result in a partially active glutamine synthetase elicit transcriptional patterns similar to those obtained by inhibition of TORC1 (Magasanik and Kaiser 2002). Similarly, treatment of cells with the glutamine synthetase-specific inhibitor, methionine sulfoximine, yields responses similar to those following treatment of cells with rapamycin (Crespo et al. 2002). However, more detailed analysis indicates that for several responses, such as Gln3 or Gat1 localization under certain conditions (see below), inhibition of glutamine synthetase has the opposite effect of that of rapamycin treatment (Tate et al. 2010). The likely conclusion is that glutamine levels provide input to the nitrogen catabolite repression pathway described above, which functions in parallel with TORC1 to effect overlapping downstream responses. Thus, the actual intracellular signal for TORC1 remains undefined but may be, as with mammalian cells, intracellular amino acid levels (Figure 4).
Saccharomyces cerevisiae regulates TORC1 using only a portion of the machinery used by mammalian cells. Saccharomyces does not encode homologs of Tsc1 or Tsc2 and its Rheb homolog is not involved in regulating TORC1. The absence of these regulatory elements may reflect elimination in yeast of input to TORC1 from growth factor receptors or AMP kinase. However, yeast TORC1 does respond to amino acid levels and the Rag family of GTP-binding proteins are retained in yeast and appear to help couple TORC1 activity to nitrogen quality and quantity, as reflected by amino acid availability.
Gtr1 and Gtr2 are yeast orthologs of RagA/B and RagC/D, respectively. These proteins, along with Meh1/Ego1 and Slm4/Ego3, form the EGO complex, which is required for microautophagy and recovery of cells from treatment with rapamycin (Dubouloz et al. 2005). Recovery from rapamycin treatment requires Gtr1 to be bound to GTP and Gtr2 to be bound to GDP, suggesting that as is the case with the mammalian Rag orthologs, the specific nucleotide binding states of Gtr1 and Gtr2 dictates function of the complex in which it acts (Binda et al. 2009). Moreover, Gtr1 locked in the GTP-bound state stimulates TORC1 in vivo and blocks the ability of cells to grow on poor nitrogen sources, which requires reduced TORC1 activity. Both genetic and biochemical evidence indicates that Gtr1, particularly when bound to GTP, physically interacts with the TORC1 components, Tco89 and Kog1, and this interaction is diminished under leucine starvation. The nucleotide binding status of Gtr1 is regulated by the Vam6 guanine nucleotide exchange factor, which is a component of the homotypic fusion and vacuole protein sorting complex in which it promotes nucleotide exchange of Ypt7, the yeast homology of mammalian Rab-7. Consistent with the biochemical role of Vam6 in Gtr1 function, vam6 mutants are defective in recovery from rapamycin treatment and exhibit reduced TORC1 activity.
Components of the TORC1 complex as well as Gtr1 and Vam6 localize predominantly to the vacuolar membrane, to the late endosome and to the intersection of those two structures. These positions remain the same regardless of whether cells are growing in nitrogen-replete medium or under leucine or nitrogen starvation. In sum, the EGO complex possesses many of the characteristics of machinery coupling amino acid levels in the cell to TORC1 activity and share many properties with the mammalian Rag complex. However, unlike the mammalian complex, regulation is not effected by EGO-dependent relocalization of TORC1 to a subcellular activation region. Rather, EGO appears to couple amino acid levels directly to TORC1 activity. The localization of the TORC1 and EGO complex to the vacuole raises the possibility that the key upstream signal for TORC1 involves mobilization of amino acids from their stores in the vacuole.
While the above model seems to account for upstream regulation of TORC1, it is likely incomplete, since gtr1 deletion strains are not defective in several TORC1-dependent cellular responses, such as transcriptional activation of nitrogen catabolite repression genes or phosphorylation control of Npr1, a protein kinase that regulates plasma membrane sorting of amino acid permeases. One alternative pathway involves a direct interaction of the cell wall integrity pathway component, Rho1, with TORC1, inducing release of Tap42 (see below) in response to various stresses, including nitrogen downshift (Yan et al. 2012). Another candidate for upstream regulation is the conserved Npr2/Npr3 complex, identified as mutants in yeast defective in induction of DAL80, a gene subject to nitrogen catabolite repression, specifically in response to nitrogen starvation (Neklesa and Davis 2009). Mutations of NPR2 or NPR3 are defective in nuclear localization of the NCR transcription factors Gat1 and Gln3 and retain Npr1 in a highly phosphorylated state in response to nitrogen starvation. These phenotypes are consistent with a model in which the Npr2/Npr3 complex inhibits TORC1 and that inhibition is alleviated by nitrogen availability, perhaps as monitored by intracellular amino acid levels. However, a direct physical link between Npr2/Npr3 and TORC1 has not been established. Given the likely existence of a second pathway working in parallel to TORC1 to effect nitrogen catabolite repression, it is not clear whether the Npr2/3 complex acts on TORC1 or on this alternative pathway.
Downstream effectors of TORC1:
Two distinct effectors—Sch9, the protein kinase B homolog discussed above, and protein phosphatase 2A—function as intermediaries between TORC1 activity and the various downstream cellular components that affect growth, metabolism, and development. The TORC1 connection to Sch9 is relatively straightforward: TORC1 directly phosphorylates Sch9 and that phosphorylation stimulates the protein kinase activity of Sch9 (Urban et al. 2007). Thus, although Sch9 requires activation by additional upstream protein kinases (see above) that perhaps provide input on other environmental conditions, TORC1 and Sch9 function as a kinase cascade connecting growth promotion to nitrogen status.
The mechanism by which PP2A transmits TORC1 activity status is less clear. TORC1 phosphorylates the essential protein Tap42, which, in its phosphorylated state forms heterodimers with the protein phosphatase 2A catalytic subunit, encoded redundantly by PPH21 and PPH22, and with the protein phosphatase 2A-like catalytic subunit, Sit4 (Di Como and Arndt 1996; Jiang and Broach 1999; Duvel et al. 2003). Pph21/22 separately forms a heterotrimeric complex with a scaffolding subunit, Tpd3, and one of two regulatory subunits, Cdc55 or Rts1, which impart different substrate specificities to the complex. Similarly, Sit4 also forms a heterodimer with one of three regulatory subunits, Sap155, Sap185, or Sap190 (Luke et al. 1996). Given the vast excess of Pph21/Pph22 and Sit4 relative to Tap42, all of these complexes likely exist concurrently within the cell. Thus, Tap42 most likely acts to direct protein phosphatase activity to specific targets rather than simply to inhibit phosphatase activity. The Tap42 interacting protein Tip41 collaborates with Tap42 in executing the phosphatase-mediated downstream functions of TORC1 (Jacinto et al. 2001; Santhanam et al. 2004; Kuepfer et al. 2007).
Analysis of biochemical studies has prompted the following working model for the role of phosphatases in TORC1 signaling (see Figure 4) (Kuepfer et al. 2007; Tate et al. 2009). Active TORC1 phosphorylates and binds Tap42 in complexes with Pph21/22 and Sit4 at the endosomal/vacuolar membrane (Yan et al. 2006). In the TORC1-bound state, these complexes remain essentially inactive due to their spatial restriction. Upon starvation or treatment with rapamycin, the complexes are released in the cytoplasm where Tap42/Tip41 directs the phosphatase activities to various downstream substrates, such as Gat1 and Mks1. The intrinsic phosphatase activity of the complexes, or other phosphatases in the cytoplasm, results in dephosphorylation of Tap42 and dissociation of the complexes with time, resulting in a self-limiting signal elicited following inactivation of TORC1.
Tap42 appears to function as a specificity factor for the catalytic phosphatase subunits, directing the phosphatases to certain substrates. For instance, rapamycin induces a Sit4- and Pph21/22-dependent dephosphorylation of the transcription factor Gln3 and Gat1 and subsequent translocation of the factors to the nucleus, where they induce transcription of NCR target genes (Beck and Hall 1999; Cardenas et al. 1999; Tate et al. 2009). Inactivation of Tap42 has no effect on NCR gene expression under normal growth conditions but significantly attenuates induction of these genes by rapamycin (Duvel et al. 2003). These results suggest that Tap42 is required for dephosphorylation of Gln3 and Gat1 following rapamycin treatment, an event catalyzed by Sit4 and Pph21/22 (Beck and Hall 1999; Tate et al. 2009). Thus, Tap42 acts in concert with phosphatase catalytic subunits to dephosphorylate downstream targets in response to rapamycin treatment, placing Tap42 as a positive regulator of phosphatase activity. Tap42 plays a similar role in rapamycin induction of RTG target genes (Duvel et al. 2003).
Phosphoproteomic studies have highlighted the bifurcation of signaling from TORC1 through Sch9 on one branch and Tap42/phosphatases on the other (Huber et al. 2009). In particular, this study examined the changes in phosphorylation following rapamycin treatment of a large number of proteins and identified changes dependent on Sch9 or Tap42. While some proteins exhibited rapamycin-induced changes that were dependent on both Sch9 and Tap42, many proteins exhibited phosphorylation changes dependent only on one or the other activity, highlighting the independence of the two downstream pathways. As noted below, the primary targets of Sch9-mediated TORC1 phosphorylation are those proteins involved in regulation of mass accumulation, including transcriptional regulators of ribosome biogenesis and ribosomal protein genes, rRNA expression and tRNA synthesis. Finally, some rapamycin-induced changes in phosphorylation occurred independently of Sch9 and Tap42, suggesting either a limit in the sensitivity of the analysis or the existence of other pathways emanating from TORC1 (Breitkreutz et al. 2010).
A second nitrogen regulatory pathway:
A variety of observations, primarily from the Cooper laboratory, have provided strong evidence that nitrogen-source regulation of NCR and RTG genes does not proceed solely through the TORC1 pathway. For instance, induction by rapamycin of the RTG responsive gene, CIT2, is nitrogen-source dependent, occurring in ammonia or glutamine but not proline or glutamate grown cells (Tate and Cooper 2003). In addition, the pattern of Gln3 phosphorylation differs in rapamycin-treated vs. nitrogen starved or methionine sulfoximine treated cells, indicating that nitrogen deprivation and rapamycin impinge on Gln3 phosphorylation status in different ways (Tate et al. 2009). Finally, Gln3 and Gat1 both regulate NCR genes but Gln3 nuclear localization occurs in response predominantly to nitrogen limitation or methionine sulfoximine treatment rather than rapamycin treatment, whereas Gat1 nuclear localization occurs in response predominantly to rapamycin treatment and is immune to nitrogen starvation or methionine sulfoximine treatment (Tate et al. 2010). These observations suggest that nitrogen availability does not regulate the TORC1 activity with regard to nitrogen response so much as provides a permissive state in which TORC1 may or may not influence nitrogen catabolite repression. For instance, Gat1 activation depends on the presence of Gln3 but Gln3 can promote transcriptional activation on its own (Georis et al. 2009). This suggests that this alternative nitrogen response pathway plays the predominant role in NCR and RTG regulation and that TORC1 inhibition can reinforce that response. As mentioned above, the nature of this alternative nitrogen regulatory pathway and the identity of its component remain unresolved. However, the Npr2/3 proteins noted above could be central players in this pathway.
The Response of Cells to Nutrient Availability
Growth control
Ribosome biogenesis:
Nutrients fuel cell growth, a process that depends primarily on the biosynthetic capacity afforded by ribosomes. Accordingly, a major role of nutrient signaling is management of ribosome biogenesis and the translational apparatus. Ribosomes comprise four RNA molecules and 79 ribosomal proteins (RPs) encoded by 138 genes. More than 236 proteins, the ribosome biogenesis (Ribi) proteins, participate in biosynthesis and assembly of the ribosome into a functional structure. tRNA molecules and a number of additional proteins are further required to produce an operational protein synthesis apparatus. Nutrient signaling impinges on biogenesis of all of these constituents, including RNA polymerase I-dependent rRNA synthesis, RNA polymerase II-dependent RP, and Ribi protein production and RNA polymerase III-dependent synthesis of tRNAs and the 5S ribosomal RNA (Figure 5).
Figure 5 .
Regulation of ribosome biogenesis. (A) Ribi gene repressors. Ribi gene expression responds to nutritional input through alleviation of repression effected by Dot1, Tod1, and Stb3, which recruit the histone deacetylase Rpd3L. Expression requires inactivation of all three repressors and, while input exhibits significant cross-talk, glucose and Ras/PKA predominantly influence Dot1 activity while nitrogen and TORC1 predominantly influence Tod6 activity. (B) rDNA, RP, and Ribi gene regulation. Regulation of rRNA transcription by nutrients involves both template activation—from a repressed, nucleosome-bound state to a locus bound predominantly by the high-mobility group protein Hmo1—as well as regulation of Pol I initiation, primarily controlled by the level and interaction of Rrn3 with Pol I. Transcriptional regulation of Ribi and ribosomal protein (RP) genes involves both local reorganization of the promoters as well as translocation of the split finger transcription factor from a cytoplasmic association with Mrs6 to the nucleus, where it peripherally associates with the promoters.
RNA polymerase I:
Nutrient availability regulates the rate of rRNA production. Three of the rRNAs, 25S, 18S, and 5.8S, are processed from a single 35S transcript encoded in 150–200 tandem repeats of the 9.1-kb rDNA gene (Johnston et al. 1997), which also encodes the RNA polymerase III transcribed 5S RNA. Four general transcription factor complexes promote transcription initiation of the 35S gene by Pol I: the upstream activation factor (UAF), core factor (CF), TATA binding protein (TBP, encoded by SPT15), and the monomeric factor Rrn3, which forms a complex with Pol I. In the current model for Pol I initiation, UAF recruits CF and TBP to the Pol I promoter, which together provide a platform for transcriptional initiation by the Rrn3–Pol I complex (Keys et al. 1996; Goetze et al. 2010). The level of the initiation competent Rrn3–Pol I complex likely serves as a key regulatory process affecting the rate of rDNA synthesis in response to nutrient availability (Grummt 2003; Claypool et al. 2004; Mayer et al. 2004; Philippi et al. 2010). Specifically, glucose depletion decreases synthesis of Rrn3 while TORC1 inhibition stimulates its proteolytic degradation and destabilizes the Rrn3–Pol I complex. Li et al. (2006) showed that Tor1 directly binds to the 35S rDNA promoter in a nutrient- and rapamycin-sensitive manner and that this physical interaction is necessary for 35S rRNA synthesis. Finally, artificially stabilizing the Rrn3–Pol I complex by physically tethering Rrn3 to the polymerase renders rDNA expression resistant to repression by nutritional downshift or rapamycin treatment (Laferte et al. 2006). In sum, these observations demonstrate that Pol I activity responds to nutritional levels via modulation of its interaction with Rrn3 (Figure 5).
Nutrient signaling also regulates RNA Pol I-dependent rRNA production at the level of template availability (Figure 5). Even under nutrient-rich conditions and rapid growth, the rDNA repeats in each cell exist in two different chromatin states, with approximately half in a transcriptionally active “open” configuration and the others in a transcriptionally inactive “closed” configuration (Conconi et al. 1989; Dammann et al. 1993). Following transition into stationary phase, the number of open repeats decreases, a process dependent on the histone deacetylase, Rpd3 (Sandmeier et al. 2002). Transcriptionally active repeats are essentially devoid of nucleosomes and instead are coated with the HMG box protein, Hmo1, while the inactive repeats are packaged in canonical nucleosomes (Merz et al. 2008; Wittner et al. 2011). The proportion of active vs. inactive repeats appears to be established by a dynamic equilibrium reset at each cell cycle. DNA replication converts most repeats into the closed, nucleosome-packaged state, a subset of which are reactivated shortly after S phase by Pol I-dependent transcription, which evicts nucleosomes and recruits Hmo1. Hmo1 maintains the open configuration for the duration of the cell cycle, even in the absence of additional transcription. Thus, the proportion of active loci likely reflects the amount of one or more limiting Pol I initiation factors responsible for transcriptional activation following S phase (Wittner et al. 2011). Accordingly, nutrient regulation of active repeat number devolves into regulation of transcriptional initiation by RNA PolI.
RNA polymerase II:
Nutritional status regulates expression of RP and Ribi genes using overlapping but distinct mechanisms (Figure 5). Ribi gene promoters possess various combinations of RRPE and PAC motifs, which serve as binding sites for Stb3 and Dot6/Tod6 factors, respectively (Jorgensen et al. 2004; Badis et al. 2008; Freckleton et al. 2009; Zhu et al. 2009). Dot6 and Tod6 function as transcriptional repressors whose repressive activities are alleviated by phosphorylation by PKA and TOR, respectively (Lippman and Broach 2009). As noted in the previous section, PKA responds to glucose availability and TOR to nitrogen availability. Thus, to a first approximation, regulation of Ribi gene expression follows a simple logic: activity of both pathways is required for elimination of both repressors; the absence of either signal results in Ribi gene repression. In other words, any single nutritional deprivation is sufficient to cause repression of ribosome biogenesis. This process is more complex in that Sch9 responds to both glucose and nitrogen availability and phosphorylates both Dot6 and Tod6, perhaps as a means of reinforcing regulation through PKA and TORC1 (Huber et al. 2009; Lippman and Broach 2009). Moreover, the nuclear localization of Stb3, which also serves primarily as a repressor of both RP and Ribi gene expression, the latter through association with the RRPE motif, is regulated by PKA and TORC1: elimination of either activity results in cytoplasmic localization of Stb3 (Liko et al. 2010). Dot6, Tod6, and Stb3 appear to effect repression of Ribi gene expression at least in part by recruitment of the Rpd3L histone deacetylatase complex to Ribi gene promoters (Humphrey et al. 2004; Huber et al. 2011).
RP gene transcription is quite sensitive to the growth potential of the cell, rapidly increasing during nutrient upshifts and rapidly decreasing during nutrient downshifts or in response to a variety of stresses. Most of the RP gene promoters bind transcriptional factors Rap1 or Abf1, which generate a nucleosome-free domain and recruit the forkhead-like transcription factor, Fhl1, which in turn recruits the transcriptional activator Ifh1 to drive gene expression (Morse 2000; Boeger et al. 2003; Lee et al. 2004; Wade et al. 2004; Zhao et al. 2006). In the absence of Ifh1, Fhl1 acts a repressor. Nutrient regulation of RP gene expression is not effected at the level of Rap1 binding. Rather, TORC1 controls RP gene expression in part by regulating the interaction between Fhl1 and Ifhl via the forkhead-associated (FHA) domain of Fhl1 (Schawalder et al. 2004; Wade et al. 2004; Rudra et al. 2005). Disruption of this domain or TORC1 inactivation by rapamycin results in loss of Ifhl from RP gene promoters and severe reduction in RP gene expression (Martin et al. 2004; Zhao et al. 2006). Moreover, although Stb3 was identified as a factor with affinity for the RRPE element in Ribi gene promoters, Stb3 associates with RP promoters in vivo and deletion of STB3 alleviates RP repression following Sch9 inactivation (Huber et al. 2011). These factors function in part by promoting chromatin remodeling. Under nutrient-replete conditions, the histone acetyl transferase, Esa1, resides at RP promoters through association with Rap1 (Reid et al. 2000; Rohde and Cardenas 2003). Under starvation conditions, Esa1 is released and the Rpd3L complex binds to the promoter, recruited predominantly by Stb3 (Huber et al. 2011).
The split finger transcription factor Sfp1 provides an additional layer of regulation of Ribi and RP genes in response to nutrients. Sfp1 is required for efficient expression of Ribi and RP genes: deletion of SFP1 results in slow growth, repression of Ribi/RP genes and very small cell volume (Jorgensen et al. 2002, 2004; Marion et al. 2004). Overexpression of Sfp1 elicits a rapid increase in Ribi gene expression and a delayed increase in RP gene expression, suggesting that the latter response is indirect. Sfp1 shows no specific binding to Ribi or RP promoter motifs in vitro and is associated only with RP promoters in vivo (Marion et al. 2004). Subcellular localization of Sfp1 is highly sensitive to nutrient status. In unstressed cells growing in rich medium, Sfp1 is located predominantly in the nucleus. Glucose or nitrogen depletion induces exodus of Sfp1 from the nucleus within minutes, a process mimicked by rapamycin treatment and blocked by hyperactivation of the PKA pathway (Jorgensen et al. 2004). Cytoplasmic Sfp1 binds to the essential Rab escort protein, Mrs6, a protein that promotes prenylation and membrane delivery of a number of constituents that control vesicle trafficking at various stages of the secretory process, particularly ER to Golgi and intra-Golgi trafficking. Sfp1–Mrs6 interaction dictates the nuclear/cytoplasmic distribution of Sfp1: increasing or decreasing Mrs6 levels leads to a corresponding increase or decrease in the fraction of Sfp1 localized to the cytoplasm (Singh and Tyers 2009). Sfp1 also binds to, and is directly phosphorylated by, TORC1 (Lempiainen et al. 2009). However, nutrient depletion does not disrupt the binding of TORC1 to Sfp1, even though (1) nutrient depletion results in eviction of Sfp1 from the nucleus and (2) rapamycin treatment disrupts the interaction. Thus, nutrients regulate Sfp1’s interaction with Mrs6. Finally, these results highlight a growing consensus that TORC1 regulatory complexes are assembled on internal membrane structures.
In short, the mechanistic basis of RP and Ribi gene regulation is still somewhat enigmatic, despite accumulation of substantial details. Regulators such as Stb3 and Sfp1 bind to one set of genes but affect expression of the other set of genes. This may indicate a cross regulation between RP and Ribi gene expression that has not been fully clarified. However, it is certainly the case that TORC1 regulates ribosome biogenesis through two parallel branches: TORC1 modulates Sch9 activity, which affects Ifh1–Fhl1 interaction at, as well as Tod6 and Stb3 binding to, Ribi gene promoters, and TORC1 influences RP gene expression by regulating Sfp1 localization. Similarly, PKA promotes Ribi gene expression through inactivation of Dot6 and Stb3 and promotes RP gene expression by stimulating Sfp1 nuclear localization through modulation of its interaction with Mrs6. Thus, carbon-source signaling primarily through PKA and nitrogen-source signaling primarily through TORC1 impinge on Ribi and RP gene regulation in a way that both activities are required for gene expression.
RNA polymerase III:
The Maf1 repressor couples expression of tRNA and other Pol III transcribed genes to nutrient availability (Willis and Moir 2007). Cells lacking Maf1 grow normally in rich media but fail to repress Pol III transcription following exit from exponential growth or following nutrient starvation or various environmental stresses. Nutrient depletion leads to relocalization of Maf1 to the nucleus where it either disengages or dislodges Pol III from transcribing genes (Oficjalska-Pham et al. 2006; Roberts et al. 2006). Signals affect Maf1 at two levels. Phosphorylation of PKA consensus sites adjacent to a nuclear localization domain on Maf1, catalyzed by either PKA or Sch9 and reversed by protein phosphatase 2A, precludes nuclear import (Oficjalska-Pham et al. 2006; Lee et al. 2009). However, nuclear localization of Maf1 is not sufficient to establish repression; phosphorylated Maf1 does not effect repression even when restricted to the nucleus (Towpik et al. 2008). Thus, phosphorylation both prevents nuclear import and blocks the ability of Maf1 to effect repression once in the nucleus. However, this model falls short of accounting for the logic of nutrient control of Pol III activity. Available data suggest that either active signaling through PKA in response to glucose or active signaling via TORC1/Sch9 in response to nitrogen should block Maf1 activity, thus requiring inactivation of both signaling pathways to effect repression. However, starvation for either nutrient alone is sufficient to elicit repression. Accordingly, some pieces of the puzzle are missing.
Metabolism:
Metabolic flux:
Central to cell growth is the metabolism of nutrients to generate energy, create building blocks for macromolecular biosynthesis, and promote synthesis of the panoply of molecules needed to make two cells from one. Yeast cells must recognize what nutrients are available and adapt their metabolic activity to match the nature and levels of those available nutrients. Several studies have documented substantial changes in the levels of various metabolites following nutrient transitions (Brauer et al. 2006; Kresnowati et al. 2006; Boer et al. 2010). For instance, phosphoenol pyruvate levels spike following glucose starvation and α-ketoglutarate levels significantly increase following nitrogen starvation. Surprisingly, the correlation between gene expression and metabolism in yeast is quite poor: metabolite levels are little influenced by the expression levels of genes encoding enzymes for their synthesis or consumption (Brauer et al. 2006; Bradley et al. 2009; Boer et al. 2010; Klosinska et al. 2011). Accordingly, metabolic flow is likely controlled by mass action combined with allosteric regulation and post-translational modification of key metabolic enzymes. The extent to which each of these processes contributes to metabolic regulation is only partially understood.
Carbon-source signaling through PKA clearly plays a role in certain metabolic processes (Figure 6). High glucose levels block synthesis and accumulation of trehalose and glycogen through PKA-dependent inhibition of the enzymes responsible for synthesis of these storage carbohydrates and activation of the enzymes responsible for their degradation (see Broach and Deschenes 1990 for references). PKA, in conjunction with protein kinase C, phosphorylates several enzymes involved in phospholipid biosynthesis, resulting in increased phosphatidyl serine production and a redirection of synthesis through the triacylglycerol biosynthetic (Kennedy) pathway relative to the diacylglycerol pathway (Carman and Han 2010). PKA signaling has also been implicated in activation of metabolic flux through the glycolytic pathway. PKA phosphorylates and activates phosphofructose kinase 2 (Dihazi et al. 2003), which synthesizes fructose-2,6 bisphosphate, an allosteric activator of phosphofructose kinase 1, a rate-limiting step in glycolysis and whose product, fructose-1,6 bisphosphate (FBP), is itself an allosteric activator of pyruvate kinase, the second rate-limiting step in glycolysis. Moreover, PKA phosphorylates pyruvate kinase directly, a modification that renders its activity less dependent on allosteric activation by FBP (Portela et al. 2002, 2006). Finally, PKA-dependent phosphorylation of the key gluconeogenic enzyme, FBP bisphosphatase, targets the enzyme for vacuolar import and degradation. Accordingly, inactivation of PKA would be expected to reduce flux through the glycolytic pathway and enhance gluconeogenesis while hyperactivation would be expected to have the opposite effect. To date, this hypothesis has not been tested. Finally, although metabolic changes attendant on nutrient transitions in snf1 and SNF1 strain have been reported (Usaite et al. 2009; Humston et al. 2011), little information is available on the possible direct effects on metabolic flux of the SNF1 signaling pathway or others, such as TORC1. Exploration of these possible connections between signaling and metabolism in yeast could prove informative, especially given the emerging awareness of the role of signaling-mediated changes in metabolic activity in diseases such as cancer (Dang et al. 2011).
Figure 6 .
Allosteric regulation of carbon metabolism. Metabolite-mediated allosteric interactions and PKA-catalyzed phosphorylations regulating metabolic flux in carbon utilization are shown in an abbreviated version of the glycolytic/gluconeogenic and storage carbohydrate pathways.
Glucose sparing:
Yeast cells starved for nitrogen, phosphate, or sulfur arrest in a quiescent state in which fermentation of glucose is suppressed: external glucose is not depleted and ethanol does not accumulate. In contrast, auxotrophs starved for the required amino acid arrest growth but continue to ferment glucose, thereby depleting external glucose and accumulating ethanol in the medium (Brauer et al. 2008). This observation suggests that cells exert cross control in metabolism such that starvation for an essential nutrient elicits a coherent growth cessation in which the cell’s metabolic activity toward other nutrients is suppressed. This cross-metabolic control is not simply the consequence of growth arrest, since “unnatural” starvation does not elicit this cross-metabolic regulation. Mutants defective in TORC1 signaling protect auxotrophs from rapid loss of viability as well as glucose wasting upon starvation for the required amino acid (Boer et al. 2008), suggesting that reduced TORC1 signaling attendant on starvation inhibits glucose metabolism.
Metabolic cycles:
Yeast cells in high-density cultures can exhibit synchronous metabolic cycles between conditions of high-oxygen consumption and low-oxygen consumption. These cycles are correlated with specific cyclical changes in expression of a large number of genes (Klevecz et al. 2004; Tu et al. 2005). The same gene expression changes can be observed in individual cells in continuous nutrient-limited chemostats where culture-wide synchrony is not ongoing, suggesting that metabolic cycling likely occurs in a cell autonomous fashion even in low-density cultures under nutrient limitation (Silverman et al. 2010). The metabolic cycle time is generally shorter than that of the cell cycle although harmonically coupled to it, such that DNA replication in any cell in the culture always occurs at the same point in the metabolic cycle but not at every metabolic cycle. The observed correlation between the specific phase of the metabolic cycle at which DNA replication occurred suggests that the metabolic cycle provided a mechanism by which DNA replication can be temporally segregated from oxidative respiration to avoid potential mutagenic effects of reactive oxygen species generated during respiration (Klevecz et al. 2004; Tu et al. 2005; Chen et al. 2007; Silverman et al. 2010). However, a recent report may suggest that this conclusion may not be universally applicable (Slavov and Botstein 2011). Moreover, the coupling between gene expression and metabolic cycling and the role of nutrient signaling in this process has yet to be resolved.
Stress response:
Yeast cells subjected to starvation for any nutrient exhibit a stereotypic pattern of gene expression changes, referred to as the environmental stress response (ESR), elicited by any of a large number of environmental insults, such as heat, oxidative stress, or high osmolarity (Gasch et al. 2000). The predominant components of the set of genes that are repressed in the ESR include those required for mass accumulation, primarily the Ribi and RP clusters described previously. That these genes are repressed following a nutrient downshift can be readily understood on the basis of their regulation by nutrient signaling pathways described above. That other stresses also elicit a similar repression suggests that the individual stressors either engage nutrient signaling pathways, such as PKA and TORC1, or interact with the same transcriptional regulatory apparatus that responds to nutrients. One potential candidate for the focal point for these convergent signals is Sfp1, which controls both Ribi and RP gene expression and whose nuclear localization responds to nutrient levels as well as to stresses such as oxidizing agents (Jorgensen et al. 2004; Singh and Tyers 2009).
Several different transcription factors are responsible for the stereotypic induction of genes in the ESR. The redundant Msn2 and Msn4 transcription factors bind to STRE elements to activate transcription of a large number of stress responsive genes. Nutrient availability impinges on Msn2/4 activity by modulating levels of phosphorylation of various sites that regulate Msn2/4 nuclear entry, nuclear exit, and transcriptional activation. Phosphorylation of Msn2/4 by either PKA or Snf1 promotes their nuclear export and blocks their nuclear import, thus preventing Msn2/4 induction of stress response genes; their dephosphorylation by PP2A and PP1 has the opposite effects (Gorner et al. 2002; De Wever et al. 2005). Inactivation of TORC1 has no direct effect on Msn2 localization, although it potentiates activation by other stresses (Santhanam et al. 2004). Finally, Yak1 phosphorylates and activates the Msn2/4 transcriptional response without affecting the localization or DNA binding of Msn2/4 (Malcher et al. 2011). Msn2/4 also responds to high osmolarity, heat stress, and oxidative agents, although the means by which these stresses are transmitted to the transcription factors are not known. The transcription factors Yap1, Hsf1, and Hog1 promote transcriptional activation in response specifically to oxidative, heat, and osmolar stresses, respectively. Except for Hsf1, which is phosphorylated by Yak1 to enhance DNA binding, these factors are generally unresponsive to nutritional input.
Deletion of MSN2 and MSN4 or deletion of YAK1, encoding a protein kinase required for full activation of Msn2 and Msn4, alleviates the growth defect from loss of PKA activity (Garrett and Broach 1989; Smith et al. 1998). Thus, surprisingly, the only essential growth-promoting activity of PKA is to attenuate the stress response mediated by Msn2/4. Why unfettered Msn2/4 activity prevents cell proliferation or survival is not clear, especially since Msn2/4 does not mediate repression of growth promoting genes such as the RP and Ribi regulons. Thus, Msn2/4 appears to provide a mechanistically undefined “brake” for cell growth, whose elimination allows growth even in the absence of the “accelerator” afforded by PKA activity.
While the ESR is robust, it does not appear to provide protection against immediate insults. Most of the genes that are induced by heat shock are not required to survive that stress (Giaever et al. 2002). Moreover, genes induced by various nutrient starvations are not required to survive those particular nutrient depletions (Klosinska et al. 2011). The fact that stress -induced genes are not required for surviving that stressful condition is not surprising, since the time lag associated with transcription and translation of new gene products would preclude a cell’s ability to mount an immediate response to stress through transcriptional activation (Tagkopoulos et al. 2008). Nonetheless, Msn2/4-dependent stress-stimulated induction of gene expression is required for an acquired stress resistance or adaptive response, in which a mild initial stress provides protection of cells against a subsequent lethal exposure to the same or a different stress (Berry and Gasch 2008). This adaptive response is not unique to yeast cells but pervades all cells in which it has been examined (Chinnusamy et al. 2004; Durrant and Dong 2004; Kensler et al. 2007; Zhao et al. 2007). Thus, cells possess intrinsic stress resistance but use transcriptional activation in response to an initial stress as a means of preparing for future stresses.
Autophagy:
Autophagy is a conserved cellular response that serves to recycle macromolecules during nutrient limitation. Yeast cells exhibit two distinct versions of autophagy—macroautophagy and microautophagy. In macroautophagy, cytoplasmic material or specific cytoplasmic organelles, such as peroxisomes, ribosomes, or mitochrondria, are packaged in a double membrane structure, termed an autophagosome, that fuses with the vacuole to deliver the cargo for degradation and recycling (Yang and Klionsky 2009). Microautophagy occurs by direct assimilation of cytoplasmic material into the vacuole through invagination of the vacuolar membrane (Uttenweiler and Mayer 2008). Both processes can be activated by starvation for any of a variety of nutrients and offer means of surviving those periods of starvation (Klosinska et al. 2011).
Autophagy is highly responsive to the nutritional status of the cells, transmitted predominantly through the TORC1 and PKA pathways (Yang and Klionsky 2009). Regulation is exerted at the level of activation of the Atg1 complex, the initial stage of autophagy that precedes nucleation of the autophagic vesicle. The Atg1 complex consists of the Atg1 protein kinase, whose activity is essential for autophagic induction, Atg13, and the Atg17–Atg31–Atg29 subcomplex. In nutrient-replete medium, TORC1 directly phosphorylates Atg13 on multiple sites, preventing Atg1 recruitment to the complex (Kamada et al. 2010). Under starvation conditions or following treatment with rapamycin, Atg13 becomes dephosphorylated, allowing association of Atg1 with the complex and activation of its protein kinase activity. Expression of a nonphosphorylatable version of Atg13 yields induction of autophagy in cells growing in rich medium, indicating that dephosphorylation of Atg13 is alone sufficient for initiating the autophagic process (Kamada et al. 2010).
The PKA pathway also influences autophagy. Activation of PKA by deletion of Bcy1, by expression of an activated RAS2 allele or by addition of cAMP, prevents induction of autophagy by rapamycin treatment or nitrogen starvation (Budovskaya et al. 2004; Yorimitsu et al. 2007). Consistent with those observations, simultaneous inactivation of PKA and Sch9 yields partial activation of the autophagic response. The locus of action of PKA and Sch9 is not entirely clear: inactivation of PKA and Sch9 does not significantly alter the phosphorylation state of Atg13. Moreover, the effect on autophagy of inactivating PKA and Sch9 is synergistic with TORC1 inactivation, suggesting that the PKA pathway acts in parallel with the TORC1 pathway. Finally, the dominant role of TORC1 over PKA in regulating autophagy may reflect the greater significance of autophagy in protecting cells from nitrogen starvation than from carbon starvation, given that the product of autophagy-induced turnover of proteins is amino acids. In fact, mutants defective in autophagy are much more sensitive to nitrogen or phosphate starvation than to glucose starvation (Klosinska et al. 2011).
Microautophagy is less extensively studied than macroautophagy and has been characterized primarily from microscopic morphological analysis and from biochemical studies (Uttenweiler and Mayer 2008). The process involves tubular invaginations into the vacuole followed by scission of the tubular structure to form vesicles within the lumen of the vacuole. It is related to piecemeal microautophagy of the nucleus, in which nuclear ER makes direct contact with the vacuole, followed by invaginations into the vacuole at the point of contact that are pinched off to generate luminal vesicles filled with nuclear material (Roberts et al. 2003; Levine and Klionsky 2004). Like macroautophagy, microautophagy is induced by nitrogen or carbon starvation or by TORC1 inactivation. The machinery for microautophagy is distinct from but overlaps that for macroautophagy (Uttenweiler and Mayer 2008; Krick et al. 2009; Dawaliby and Mayer 2010). In addition to its role in the starvation response, microautophagy serves to restore cell growth following rapamycin treatment, at least in part by reducing the amount of vacuolar membrane that accumulates as a result of starvation-induced macroautophagy. This process requires the previously described EGO complex, comprising the small G proteins, Gtr1 and Gtr2, along with Meh1/Ego1 and Slm4/Ego3 (Dubouloz et al. 2005; Binda et al. 2009), which is also required for activation of TORC1 by amino acids. In fact, the effect of the EGO complex on microautophagy may result solely from its function as an activator of TORC1 activity.
Development
In addition to controlling cell growth, nutrients dictate which of a number of developmental programs a cell chooses to pursue. For instance, under nutrient-limiting conditions cells can engage in pseudohyphal growth and invasion of the substratum, perhaps as a method of “foraging” for nutrients. Haploid cells starved for any essential nutrient can exit the mitotic cycle and assume a nutrient-specific quiescent state, which allows extended survival under the particular starvation condition. Finally, diploid cells subject to substantial starvation can undergo meiosis and sporulation. The spores that emerge from this process are capable of weathering extreme conditions for extended periods of time. The particular developmental program pursued is dictated by a complex interplay of the signaling networks responsive to nutrient availability and to stress. The specific developmental pathways are discussed in more detail in other chapters in this volume. Accordingly, the following focuses primarily on nutrient regulation of those processes.
Filamentous growth:
Diploid cells subjected to limiting nitrogen or haploid cells subjected to limiting glucose become elongated, exhibit polar budding, suppress budding in mother cells, remain attached after cytokinesis, and elaborate extracellular glucanases (Gancedo 2001; Palecek et al. 2002). This ensemble of features yields chains of cells, referred to as pseudohyphae in diploids or filaments in haploids, capable of invading the substratum. Depending on the nature of the strain and whether the cells are in liquid or on a solid surface, such cells could exhibit other behaviors, including biofilm formation, flocculation, or surface flotation (Bruckner and Mosch 2011). While these programs have been viewed as distinct processes responding to different nutritional cues, recent studies have shown that the signaling pathways responsible for all these programs overlap significantly. Nutrient regulation of filamentous growth impinges on many cellular processes and affects a large number of genes. This regulation has been reviewed recently (Zaman et al. 2008; Bruckner and Mosch 2011), so I simply highlight a few of the more recent observations, focusing primarily on the regulation of FLO11, a major adhesion gene whose expression is tightly associated with filamentation.
The transcriptional changes associated with filamentation are coordinated by a collection of transcription factors responsive to the various nutrient signaling pathways described in the previous sections (Figure 7). These factors include the transcriptional activators Ste12, Tec1, Ash1, Flo8, Phd1, Mss11, Msn1, Haa1, and Mga1 and the transcriptional repressors Sok2, Nrg1, Nrg2, and Sfl1. These factors comprise a highly connected and complex network with Mga1 and Phd1 serving as the predominant modulators of the filamentous transitions (Borneman et al. 2006). These transcription factors mediate expression of several hundred genes that change expression during transition from yeast to pseudohyphal growth (Prinz et al. 2004). In some cases, these factors all converge on the promoter of a gene, such as FLO11, whose expression is required for pseudohyphal growth but more often these factors regulate distinct but overlapping suites of genes whose concerted action orchestrates the filamentous program.
Figure 7 .
Nutrient regulation of Flo11 expression. The large FLO11 promoter exists in three states: epigenetically silenced (lower), permissive for activation (middle), and activated (upper). Transition between the silenced and permissive state occurs slowly and is associated with the alternative binding of the Sfl1 repressor and the Flo8 activator, which regulate expression of two upstream noncoding RNAs, the antisense PWR1 transcript and the sense ICR1 transcript. Extended transcription of ICR1 interferes with activation from the FLO11 promoter, associated with nucleosome-mediated occlusion of the transcriptional start site. In the permissive state, in which PWR1 expression blocks extension of ICR1 into the promoter, various activators (blue) and repressors (red) modulate expression of the gene in response to environmental conditions via various signaling networks (gray).
Carbon source regulation of filamentation and the signaling pathways mediating that regulation is confounding. Haploid invasiveness and diploid filamentation are stimulated by carbon starvation through the Snf1 pathway, which inactivates the transcriptional repressors, Nrg1 and Nrg2. However, stimulation of the Ras/PKA pathway, by an activated RAS2 allele, an activated GPA2 allele, inactivation of Ira1, or addition of cAMP, also stimulates filamentation, even though enhanced PKA activity is associated with high glucose levels. One possible explanation for this discrepancy is that PKA regulation of filamentation may not be related to its function in glucose signaling, which is realized on a relatively short time scale, but rather to a long time-scale role for PKA, perhaps in cell–cell communication on the basis of diffusion of metabolic byproducts, not unlike quorum sensing in bacteria (Chen and Fink 2006; Wuster and Babu 2010). The role of PKA in filamenation has an added complexity in that Tpk2 stimulates FLO11 expression by activating the Flo8 transcriptional activator but Tpk1 inhibits FLO11 expression by inhibiting Yak1 kinase, which allows Sok2 to inhibit, and prevents Phd1 from activating, FLO11 transcription. Thus, activation of PKA sends both positive and negative regulatory signals, at least for adhesion (Robertson and Fink 1998; Malcher et al. 2011), perhaps allowing fine tuning of the adhesive response under a variety of conditions.
As discussed previously, several signaling systems influence diploid filamentation in response to nitrogen limitation, although no coherent model for nitrogen sensing emerges from these anecdotal observations. Mutations in a number of genes involved in nitrogen regulation and assimilation affect filamentation in response to nitrogen limitation. These include genes encoding the high-affinity Mep2 ammonia permease, the regulators—Gln3 and Ure2—of the NCR, those involved in amino acid permease induction, and one of the glutamine tRNA genes. However, rapamycin treatment inhibits rather than stimulates filamentation, even at sublethal doses, suggesting at most an indirect role of TORC1 in filamenation. Finally, Snf1 T210 phosphorylation is stimulated not only by glucose limitation but also by nitrogen limitation, even in the presence of high levels of glucose (Orlova et al. 2006, 2010). Thus, Snf1 activation provides the most consistent connection between filamentation and the multiple forms of nutritional deprivation required for filamentation.
Filamentation is also subject to epigenetic regulation (Figure 7). Even under conditions promoting filamentation, the FLO11 gene is persistently inactive in a subset of cells in the population while active in others (Halme et al. 2004). Inactivation of HDA1, encoding one of the yeast histone deacetylases, results in uniformly high level FLO11 expression in all cells, indicating that the inactive state of the gene results from a heritable, chromatin-based repression, similar to telomere position effect. The regulation of FLO11 expression can best be appreciated by postulating that the FLO11 promoter can exist in three distinct states—a closed or silenced state, an open competent state, and a transcriptionally engaged state—and that the regulatory factors influence transitions between those states (Octavio et al. 2009). The silenced state is impervious to transcriptional activation but can undergo an infrequent transition into a competent state, which, while transcriptionally inactive, can be rapidly converted to the active state by the appropriate combination of transcription factors. Some transcription factors influence the transition between silence and competence, while others serve only to stimulate transcription of competent promoters, while other transcription factors can perform both functions. Moreover, this transition is influenced by two upstream noncoding RNAs, whose expression responds to Rpd3L, an observation that accounts for the unexpected role of a histone deacetylase in activating gene expression (Bumgarner et al. 2009). In short, like rRNA expression, chromatin structure dictates transcription factor accessibility to the template while the signaling pathways can both influence the distribution of open vs. closed templates and the transcriptional activity of the open templates.
Quiescence:
Yeast cells, like all other living cells, spend most of their time in a quiescent state, which in yeast results from starvation for one or more nutrients. Haploid or diploid yeast cells starved for carbon, nitrogen, phosphate, or sulfur cease accumulating mass, arrest cell cycle progression prior to “start,” and enter the poorly defined G0 state. Investigators have ascribed to G0 yeast cells a number of distinguishing characteristics, including a thickened cell wall; increased storage carbohydrates; compacted chromatin; substantially reduced translation; a specific transcriptional profile; and enhanced resistance to heat, oxidative stress, and high osmolarity (Gray et al. 2004; Smets et al. 2010). However, the sole unequivocal trait of quiescence is the ability to maintain viability when starved and to resume growth following restoration of the missing nutrient. This characteristic distinguishes cells suffering from “natural” starvation, for a carbon source, for instance, from those subject to an unnatural starvation, such as auxotrophic cells deprived of the required amino acid. In the latter case, cells rapidly lose viability even though they arrest uniformly as unbudded cells (Saldanha et al. 2004; Boer et al. 2008; Brauer et al. 2008). Thus, the quiescent G0 state requires the coordinated and deliberate adaptation of cells to depletion of a core nutrient and not simply the cessation of growth that attends abrogation of protein or RNA synthesis.
The nature of quiescence and the role of signaling pathways in orchestrating quiescence have been described in several excellent reviews (Gray et al. 2004; Smets et al. 2010; De Virgilio 2011). However, these reviews focused on stationary phase cells. Recent studies have shown that many of the attributes ascribed to stationary phase cells, such as resistance to various stresses and thickened cell walls, are simply extensions of those of slow growing cells (Lu et al. 2009; Klosinska et al. 2011). Moreover, a number of attributes of stationary phase cells, such as formation of actin bodies or aggregates of a number of other proteins, are not observed in cells starved for other nutrients (Sagot et al. 2006; Narayanaswamy et al. 2009). Thus, different starvations appear to yield different quiescent states and characteristics previously ascribed to quiescent cells do not uniquely define them. In the following, I describe the features that are common to, as well as those that are distinct among, different quiescent cells and discuss the role of nutrient signaling pathways in achieving those features.
All quiescent cells are capable of extended survival under starvation conditions, although the means by which that is accomplished depends on the specific conditions. Cells starved for carbon, nitrogen, or phosphate alter their transcriptional profile, with roughly equal contributions from each of three classes of genes: those whose expressions are simply extrapolations of a growth-rate–dependent response, those that respond to a specific starvation, and those that are quiescent specific, independent of growth rate or specific nutrient starvation (Klosinska et al. 2011). Thus, many genes change expression following a specific starvation, but only a subset of those genes comprise a core quiescence transcriptional program. However, none of the genes comprising the core transcriptional profile are required for surviving quiescence. Moreover, it is not clear whether the transcripts that accumulate specifically during quiescence are translated during that time or rather are stored in P bodies or stress granules to be activated when needed at a later time, such as during restoration of growth upon refeeding (Aragon et al. 2006; Parker and Sheth 2007; Arribere et al. 2011; Ramachandran et al. 2011). Thus, transcriptional profiling does not offer significant insight into the means by which cells establish a quiescence program and survive extended nutrient depletion.
Genetic screens have identified different classes of genes required for surviving different starvations. Mutants defective in mitochondrial function rapidly lose viability upon glucose or phosphate starvation but exhibit normal survival upon nitrogen starvation. Mutants defective in autophagy are sensitive to nitrogen or phosphate starvation but survive normally on glucose starvation (Lavoie and Whiteway 2008; Gresham et al. 2011; Klosinska et al. 2011). These patterns suggest that survival requires the ability to utilize intracellular resources to mitigate the consequences of losing a particular nutrient. Consistent with this interpretation, nutrient signaling pathways are critical for surviving the specific nutrient sensed by the signaling pathway. Both snf1 and ira2 mutants, which fail to adequately inform the cell of diminished glucose levels, are sensitive to glucose starvation but not nitrogen or phosphate starvation (Klosinska et al. 2011). Similarly, rim15 mutants, a downstream target of TORC1 required for entry into stationary phase and meiosis, are particularly sensitive to nitrogen starvation (Pedruzzi et al. 2003; Klosinska et al. 2011). Thus, appropriate perception of nutrient limitation is critical for a cell to mount an appropriate quiescent program.
While nutrient-specific pathways inform cells regarding specific nutrient deprivations, cross-talk between different nutrient sensing pathways can allow one pathway to impinge on the response to a different input. As a consequence, hyperactivity of one nutrient signaling pathway can preclude attainment of quiescence under other nutrient limitation. For instance, bcy1 mutants, which exhibit PKA hyperactivation, are exquisitely sensitive to starvation for any nutrient. In a reciprocal fashion, down regulation of one signaling pathway can promote survival upon growth arrest from an unrelated starvation. For instance, any of a number of mutations diminishing TORC1 signaling protects cells from death attendant on growth arrest following depletion of an auxotroph for its required amino acid. Such mutants also exhibit extended survival in stationary phase (Powers et al. 2006; Boer et al. 2008). These observations suggest some overlap among programs that promote survival upon nutrient deprivation, both through convergence on common effectors as well as through direct cross-talk (De Virgilio 2011).
The role of signaling pathways in survival of cells in quiescence is not simply transmitting a sufficiently low signal necessary to induce entry into quiescence. Successful survival of starvation requires three distinct steps: induction of the quiescent state, maintenance of viability during quiescence, and reentry into mitotic growth upon restoration of the missing nutrient (Gray et al. 2004). Nutrient signaling pathways are critical not only for the initial stage of eliciting the quiescent state but also for exiting quiescence. Strains carrying hypoactive ras2 alleles exhibit a delay in recovery from glucose starvation (Jiang et al. 1998). Similarly, tco89 and tor1 mutants are sensitive to nitrogen starvation, likely due to a failure to reemerge from quiescence (Klosinska et al. 2011). In the same vein, mutants in the Ego complex, required for TORC1 stimulation, are defective in recovery from rapamycin-induced growth arrest (Dubouloz et al. 2005). Thus, nutrient signaling pathways participate in both entry and exit from quiescence and, accordingly, must remain responsive to the nutritional environment.
In sum, yeast cells can attain distinct quiescent states, each of which allows survival under the condition of a particular nutrient deprivation. How cells survive those deprivations is still unclear. It is likely that the most informative mutants regarding quiescence have not been identified, since such mutants would likely be lethal as they could not survive storage. In fact, bcy1 mutants cannot be stored without acquiring suppressor mutations. Thus, further genetic analysis of quiescence, focusing on essential genes, could inform how cells survive inhospitable conditions and extend lifespan in the face of adversity.
Meiosis:
MATa/MATα diploid cells that are respiratory sufficient can exit the mitotic cycle and initiate a developmental program leading to meiosis and sporulation in response to a nutritional environment that meets three criteria: the absence of one essential growth nutrient such that cells arrest in G1; the absence of glucose; and the presence of a nonfermentable carbon source (Kupiec et al. 1997; Honigberg and Purnapatre 2003; Piekarska et al. 2010). A nonfermentable carbon source is required only through premeiotic DNA replication but respiration is required throughout meiosis (Jambhekar and Amon 2008). Since autophagy mutants fail to sporulate, subsequent stages of meiosis are likely fueled by respiratory catabolism of internal stores, accounting for the requirement for continued respiration.
Initiation of meiosis requires activation of a set of early meiotic genes (EMGs) that elicit early meiotic events, such as premeiotic DNA replication, as well as precipitate the subsequent transcriptional cascade promoting middle and late meiotic gene expression. Under mitotic growth conditions, expression of a key meiotic regulator, IME2, as well as other EMGs, is repressed by Ume6-mediated recruitment of the histone deacetylases Sin3 and Rpd3 and the chromatin remodeling complex Isw2 to the URS1 sequence in the promoters of the regulated genes. Under meiotic induction conditions, Sin3 and Rpd1 dissociate from Ume6, alleviating repression, while Ime1, the primary initiator of meiosis, associates with Ume6 to recruit the histone acetyl transferase Gcn5 and activate transcription. This induction of early stage meiosis-specific genes is relatively linear in response to stimuli. Moreover, this stage of meiosis exhibits the most cell-to-cell variability in duration, with the length of this period related both to the size of the cell at onset of starvation and the overall levels of accumulation of Ime1 (Nachman et al. 2007). This variation may afford cells the opportunity to hedge their bets with regard to commitment to an irreversible developmental program, allowing some cells to remain capable of returning to growth if conditions improve even after other cells have completed the sporulation program. Once the level of Ime1 reaches a critical level, cells become irreversibly committed to meiosis and sporulation and the subsequent stages of meiosis unfurl in a temporally precise manner, with expression of the middle and late genes exhibiting a cooperative behavior (Gurevich and Kassir 2010).
Nutrient signals impinge on two key regulators that activate early meiotic gene expression—the transcription factor Ime1 and the protein kinase Ime2 (Figure 8; reviewed in Kupiec et al. 1997; Honigberg and Purnapatre 2003; Zaman et al. 2008; Piekarska et al. 2010). Most of the environmental signals converge on the large promoter of the IME1 gene, whose product activates transcription of IME2 and a host of EMGs. The products of the MATa and MATα mating-type genes comprise a heterodimeric repressor complex that inhibits transcription of the regulator of meiosis, Rme1, which otherwise binds to and represses transcription of IME1 through the USC3 and USC4 domains of the promoter. In this way, diploidy relieves one of the repressive components of IME1 expression.
Figure 8 .
Nutrient regulation of meiosis. Nutrients control entry into meiosis through regulation of IME1 transcription (left) and Ime1 function (right) in induction of IME2 and a number of early meiotic genes (EMGs). Alkaline pH resulting from oxidation of acetate activates Rim101 by proteolytic cleavage to inactivate the IME1 repressor, Smp1. Absence of glucose reduces PKA activity, leading to inactivation of the Sok2 repressor and activation of the Msn2,4 transcriptional activators. The absence of PKA also permits activation of the Rim11 and Rim15 kinases, which influence Ime1 function. Phosphate or nitrogen deprivation reduces cyclin dependent kinase (CDK) activity, permitting entry of new translated Ime1 into the nucleus. Nitrogen and phosphate, through TORC1 and Pho80/Pho85 kinases, respectively, affect access of the Rim15 kinase to the nucleus.
Glucose influences IME1 expression primarily through the PKA pathway. PKA phosphorylates and inhibits Msn2 and Msn4, which serve as transcriptional activators of IME1 through a stress-responsive IRE element in the promoter, and PKA activates the Sok2 repressor, which inhibits transcription of IME1 through binding at or near the Msn2/4 binding site. Glucose through PKA also affects the function of Ime1 by promoting phosphorylation and inactivation of Rim11 and Rim15, two protein kinases that stimulate interaction of Ime1 with the transcriptional complexes at EMG promoters. Thus, glucose affects both expression of IME1, by activating a repressor and inactivating an activator of the gene, and the subsequent function of Ime1 in its role as a transcriptional activator.
Nitrogen starvation is the normal laboratory condition for sporulation, although starvation for phosphate or sulfur can also induce sporulation even in the presence of an adequate nitrogen source. The primary regulatory role of starvation in eliciting meiosis and sporulation is to lower the level of Cln/Cdk activity. However, nitrogen starvation or TORC1 inhibition may also play an ancillary role in regulation of IME1 transcriptional initiation, by regulating Rim15 localization, for example (Swinnen et al. 2006).
The GSK3-β homology Rim11 and the stationary phase protein kinase Rim15 stimulate expression of EMGs in response to a variety of nutritional inputs. Both Rim11 and Rim15 are inactivated by phosphorylation by PKA and so are active only in the absence of PKA signaling (Rubin-Bejerano et al. 2004; Swinnen et al. 2006). In addition, TORC1 blocks nuclear import of Rim15 while Pho80/Pho85 in response to exogenous phosphate stimulates nuclear export of Rim15. Rim15, perhaps through phosphorylation of Ume6, promotes dissociation of Sin3 and Rpd3, while Rim11 phosphorylation of Ime1 stimulates interaction of Ime1 with Ume6 (Pnueli et al. 2004). Finally, Cln/Cdk blocks nuclear import of Ime1, an impediment to EMG activation that is alleviated by nutrient-starvation–induced arrest in G1. Thus, nutritional cues inform initiation of meiosis through a variety of routes.
The presence of a nonfermentable carbon source is perceived by the cell as a consequence of its metabolism to CO2 and resultant alkalization of the media. High external pH activates a highly conserved pH sensing pathway comprising cell surface receptors and the Rim101 transcription factor (formerly Rim1), which is activated by proteolytic cleavage catalyzed by the Rim13 protease. Rim101 regulates initiation of meiosis and adaptation to external alkalization by repressing the transcriptional repressors Smp1 and Nrg1 (Lamb and Mitchell 2003). The effect of Rim101 on IME1 expression is mediated by a UASrm site in the promoter, the likely binding site for Smp1 and/or Nrg1, whose repressive activity would be alleviated by Rim101. The fact that deletion of SMP1, but not NRG1, alleviates the sporulation defect in rim101Δ strains would argue that Smp1 is the immediate regulator of IME1 (Lamb and Mitchell 2003). Finally, glucose also likely influences this signaling pathway through repression of respiration, thereby blocking alkalization.
As noted above, addition of nutrients to cells undergoing meiosis can abrogate the developmental program and restore cells to mitotic growth. The targets for this nutritional control of meiotic progression have not been fully defined, although some information has emerged. For instance, Ime2, which is required at multiple stages during meiotic progression, is destabilized by glucose addition through degradation targeted by the SCFGrr1 ubiquitin ligase (Purnapatre et al. 2005). However, while expression of a degradation-resistant version of Ime2 renders cells resistant to the glucose-induced block to meiotic DNA replication, it does not render later steps in meiosis resistant to glucose. Thus, additional nutritionally sensitive processes have yet to be identified.
Conclusions and Prospectives
We have a detailed understanding of some of the circuitry underlying nutritional sensing in yeast, but we are still somewhat vague on others. For instance, the interplay of positive and negative regulators and the various feed-forward and feed-back loops in regulating expression of glucose transporters is so well described that modeling efforts have yielded highly predictive dynamic descriptions of its behavior. On the other hand, we still have no clear understanding of the upstream components of glucose signaling regulating protein kinase A or the interplay between the various small G proteins in that process. Even more poorly described are the pathways sensing and responding to nitrogen levels. While many of the components of the TORC1 signaling network have been identified and their interactions defined, we have less understanding of the pathways emanating from TORC1, particularly through protein phosphatases. Moreover, we can infer the existence of a second nitrogen-sensing pathway from the limits of TORC1 effects, but this pathway is poorly defined. Finally, we appreciate that significant cross-talk exists between and among the various nutrient signaling pathways—for instance, glucose sparing in nitrogen- or phosphate-starved cells—but the nature of that interplay is undefined. Thus, we have a number of important details to fill in regarding the structure of the nutrient-sensing networks. In addition, several fundamental questions regarding the interplay of nutrient availability and growth have yet to be solved.
Key unanswered questions
How is cell growth controlled in response to nutrients? Cells can adapt their growth rate over at least a 10-fold range in response to limiting nutrients and diminished growth rate is associated with downregulation of biosynthetic capacity and upregulation of stress response genes. What is cause and effect in this growth control? Do limiting nutrients result in limiting metabolic capacity, which is sensed through reduced energy charge or the inability to synthesize key components required for continued biomass accumulation? Or, do nutrient levels through signaling pathways set the biosynthetic, metabolic, and transcriptional program appropriate for the perceived levels of nutrients. This question is fundamental to understanding regulation of cell growth in any organism.
How have evolutionary pressures shaped the growth capacity of yeast cells? Most of the studies on yeast growth and metabolism have been conducted in nutrient-replete conditions with cells undergoing exponential growth. However, yeast cells in the wild seldom experience such a lush environment but rather struggle under nutrient-limited or -depleted conditions. Accordingly, survival and evolution likely depended more on the ability of cells to withstand adverse conditions, through developmental programs such as filamentation, quiescence, and sporulation, than on the ability to grow rapidly under nutrient-replete conditions. Survival under these conditions also requires that cells be capable of responding effectively to changing nutritional status by rapidly sensing those changes and transitioning from one state to another. Moreover, cells have survived in a highly variable and uncertain environment. Mounting a response that allows cells to “hunker down” in response to stress may be the appropriate behavior if the stress continues but would be disadvantageous if conditions improved and the stress response precluded cells from rapidly taking advantage of the improved conditions. Thus, cell populations that exhibit a diverse set of behaviors to a given environmental stress may be better positioned to survive a variable environment than those that mount uniform responses. All these issues suggest that understanding the biology of yeast, considering the pressures that shaped its biology, require exploring the ways yeast cells respond to challenging conditions and the means by which they make transitions between different nutrient states. These areas offer rich topics for future research.
Acknowledgments
Work from my laboratory described in this review was supported by National Institutes of Health (NIH) grant RO1 GM076562 with additional support from a Center for Quantitative Biology/NIH grant P50 GM071508.
Footnotes
Communicating editor: A. Hopper
Literature Cited
- Ahuatzi D., Herrero P., de la Cera T., Moreno F., 2004. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 279: 14440–14446 [DOI] [PubMed] [Google Scholar]
- Ahuatzi D., Riera A., Pelaez R., Herrero P., Moreno F., 2007. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem. 282: 4485–4493 [DOI] [PubMed] [Google Scholar]
- Aragon A. D., Quinones G. A., Thomas E. V., Roy S., Werner-Washburne M., 2006. Release of extraction-resistant mRNA in stationary phase Saccharomyces cerevisiae produces a massive increase in transcript abundance in response to stress. Genome Biol. 7: R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Doudna J. A., Gilbert W. V., 2011. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol. Cell 44: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K., Lin S. S., Manchester J. K., Gordon J. I., 2000. Sip2p and its partner snf1p kinase affect aging in S. cerevisiae. Genes Dev. 14: 1872–1885 [PMC free article] [PubMed] [Google Scholar]
- Badis G., Chan E. T., van Bakel H., Pena-Castillo L., Tillo D., et al. , 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn Y. S., Xue C., Idnurm A., Rutherford J. C., Heitman J., et al. , 2007. Sensing the environment: lessons from fungi. Nat. Rev. Microbiol. 5: 57–69 [DOI] [PubMed] [Google Scholar]
- Beck T., Hall M. N., 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692 [DOI] [PubMed] [Google Scholar]
- Belinchon M. M., Gancedo J. M., 2007. Glucose controls multiple processes in Saccharomyces cerevisiae through diverse combinations of signaling pathways. FEMS Yeast Res. 7: 808–818 [DOI] [PubMed] [Google Scholar]
- Berry D. B., Gasch A. P., 2008. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell 19: 4580–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram P. G., Choi J. H., Carvalho J., Ai W., Zeng C., et al. , 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275: 35727–35733 [DOI] [PubMed] [Google Scholar]
- Binda M., Peli-Gulli M. P., Bonfils G., Panchaud N., Urban J., et al. , 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35: 563–573 [DOI] [PubMed] [Google Scholar]
- Boeger H., Griesenbeck J., Strattan J. S., Kornberg R. D., 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11: 1587–1598 [DOI] [PubMed] [Google Scholar]
- Boer V. M., Amini S., Botstein D., 2008. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc. Natl. Acad. Sci. USA 105: 6930–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer V. M., Crutchfield C. A., Bradley P. H., Botstein D., Rabinowitz J. D., 2010. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol. Biol. Cell 21: 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Leigh-Bell J. A., Yu H., Bertone P., Gerstein M., et al. , 2006. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 20: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P. H., Brauer M. J., Rabinowitz J. D., Troyanskaya O. G., 2009. Coordinated concentration changes of transcripts and metabolites in Saccharomyces cerevisiae. PLOS Comput. Biol. 5: e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Saldanha A. J., Dolinski K., Botstein D., 2005. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell 16: 2503–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Yuan J., Bennett B. D., Lu W., Kimball E., et al. , 2006. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. USA 103: 19302–19307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Huttenhower C., Airoldi E. M., Rosenstein R., Matese J. C., et al. , 2008. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell 19: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., et al. , 2010. A global protein kinase and phosphatase interaction network in yeast. Science 328: 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Deschenes R. J., 1990. The function of ras genes in Saccharomyces cerevisiae. Adv. Cancer Res. 54: 79–139 [DOI] [PubMed] [Google Scholar]
- Bruckner S., Mosch H. U., 2011. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 36: 25–58 [DOI] [PubMed] [Google Scholar]
- Budhwar R., Lu A., Hirsch J. P., 2010. Nutrient control of yeast PKA activity involves opposing effects on phosphorylation of the Bcy1 regulatory subunit. Mol. Biol. Cell 21: 3749–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Reggiori F., Klionsky D. J., Herman P. K., 2004. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279: 20663–20671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Stephan J. S., Deminoff S. J., Herman P. K., 2005. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 102: 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner S. L., Dowell R. D., Grisafi P., Gifford D. K., Fink G. R., 2009. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. USA 106: 18321–18326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M. E., Cutler N. S., Lorenz M. C., Di Como C. J., Heitman J., 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13: 3271–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Han G. S., 2010. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80: 859–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Fink G. R., 2006. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 20: 1150–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Jiao Z. H., Zheng L. S., Zhang Y. Y., Xie S. T., et al. , 2009. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature 459: 1146–1149 [DOI] [PubMed] [Google Scholar]
- Chen Z., Odstrcil E. A., Tu B. P., McKnight S. L., 2007. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science 316: 1916–1919 [DOI] [PubMed] [Google Scholar]
- Cherkasova V., Qiu H., Hinnebusch A. G., 2010. Snf1 promotes phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol. Cell. Biol. 30: 2862–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Schumaker K., Zhu J. K., 2004. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Claypool J. A., French S. L., Johzuka K., Eliason K., Vu L., et al. , 2004. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell 15: 946–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S., Ma P., Cauwenberg L., Winderickx J., Crauwels M., et al. , 1998. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17: 3326–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Widmer R. M., Koller T., Sogo J. M., 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57: 753–761 [DOI] [PubMed] [Google Scholar]
- Cooper T. G., 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26: 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo J. L., Powers T., Fowler B., Hall M. N., 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99: 6784–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R., Lucchini R., Koller T., Sogo J. M., 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21: 2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., Hamaker M., Sun P., Le A., Gao P., 2011. Therapeutic targeting of cancer cell metabolism. J. Mol. Med. 89: 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawaliby R., Mayer A., 2010. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions. Mol. Biol. Cell 21: 4173–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C., 2011. The essence of yeast quiescence. FEMS Microbiol. Rev. 36: 306–339 [DOI] [PubMed] [Google Scholar]
- De Virgilio C., Loewith R., 2006a. The TOR signalling network from yeast to man. Int. J. Biochem. Cell Biol. 38: 1476–1481 [DOI] [PubMed] [Google Scholar]
- De Virgilio C., Loewith R., 2006b. Cell growth control: little eukaryotes make big contributions. Oncogene 25: 6392–6415 [DOI] [PubMed] [Google Scholar]
- De Wever V., Reiter W., Ballarini A., Ammerer G., Brocard C., 2005. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 24: 4115–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R., Binda M., Lee S. S., Pelet S., Winderickx J., et al. , 2010. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 29: 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J. L., Iyer V. R., Brown P. O., 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686 [DOI] [PubMed] [Google Scholar]
- Di Como C. J., Arndt K. T., 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10: 1904–1916 [DOI] [PubMed] [Google Scholar]
- Dihazi H., Kessler R., Eschrich K., 2003. Glucose-induced stimulation of the Ras-cAMP pathway in yeast leads to multiple phosphorylations and activation of 6-phosphofructo-2-kinase. Biochemistry 42: 6275–6282 [DOI] [PubMed] [Google Scholar]
- Dilova I., Aronova S., Chen J. C., Powers T., 2004. Tor signaling and nutrient-based signals converge on Mks1p phosphorylation to regulate expression of Rtg1.Rtg3p-dependent target genes. J. Biol. Chem. 279: 46527–46535 [DOI] [PubMed] [Google Scholar]
- Dombek K. M., Kacherovsky N., Young E. T., 2004. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J. Biol. Chem. 279: 39165–39174 [DOI] [PubMed] [Google Scholar]
- Dong X., Mitchell D. A., Lobo S., Zhao L., Bartels D. J., et al. , 2003. Palmitoylation and plasma membrane localization of Ras2p by a nonclassical trafficking pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F., Deloche O., Wanke V., Cameroni E., De Virgilio C., 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19: 15–26 [DOI] [PubMed] [Google Scholar]
- Durrant W. E., Dong X., 2004. Systemic acquired resistance. Annu. Rev. Phytopathol. 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Duvel K., Santhanam A., Garrett S., Schneper L., Broach J. R., 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11: 1467–1478 [DOI] [PubMed] [Google Scholar]
- Flick K. M., Spielewoy N., Kalashnikova T. I., Guaderrama M., Zhu Q., et al. , 2003. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 14: 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L., 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3: 1166–1178 [DOI] [PubMed] [Google Scholar]
- Freckleton G., Lippman S. I., Broach J. R., Tavazoie S., 2009. Microarray profiling of phage-display selections for rapid mapping of transcription factor-DNA interactions. PLoS Genet. 5: e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadura N., Robinson L. C., Michels C. A., 2006. Glc7-Reg1 phosphatase signals to Yck1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics 172: 1427–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M., 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25: 107–123 [DOI] [PubMed] [Google Scholar]
- Garrett S., Broach J., 1989. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 3: 1336–1348 [DOI] [PubMed] [Google Scholar]
- Garrett S., Menold M. M., Broach J. R., 1991. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol. Cell. Biol. 11: 4045–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georis I., Feller A., Tate J. J., Cooper T. G., Dubois E., 2009. Nitrogen catabolite repression-sensitive transcription as a readout of Tor pathway regulation: the genetic background, reporter gene and GATA factor assayed determine the outcomes. Genetics 181: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Goetze H., Wittner M., Hamperl S., Hondele M., Merz K., et al. , 2010. Alternative chromatin structures of the 35S rRNA genes in Saccharomyces cerevisiae provide a molecular basis for the selective recruitment of RNA polymerases I and II. Mol. Cell. Biol. 30: 2028–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Martinez-Pastor M. T., Estruch F., Ammerer G., et al. , 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Wolf J., Brown E. L., Ammerer G., et al. , 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., et al. , 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68: 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D., Boer V. M., Caudy A., Ziv N., Brandt N. J., et al. , 2011. System-level analysis of genes and functions affecting survival during nutrient starvation in Saccharomyces cerevisiae. Genetics 187: 299–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffioen G., Branduardi P., Ballarini A., Anghileri P., Norbeck J., et al. , 2001. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol. Cell. Biol. 21: 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., 2003. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17: 1691–1702 [DOI] [PubMed] [Google Scholar]
- Gurevich V., Kassir Y., 2010. A switch from a gradient to a threshold mode in the regulation of a transcriptional cascade promotes robust execution of meiosis in budding yeast. PLoS ONE 5: e11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J. S., Thiele D. J., 2004. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J. Biol. Chem. 279: 5169–5176 [DOI] [PubMed] [Google Scholar]
- Halme A., Bumgarner S., Styles C., Fink G. R., 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415 [DOI] [PubMed] [Google Scholar]
- Harashima T., Heitman J., 2005. Galpha subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol. Biol. Cell 16: 4557–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima T., Anderson S., Yates J. R., III, Heitman J., 2006. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol. Cell 22: 819–830 [DOI] [PubMed] [Google Scholar]
- Hardie D. G., Carling D., Carlson M., 1998. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67: 821–855 [DOI] [PubMed] [Google Scholar]
- Hardie D. G., Carling D., Gamblin S. J., 2011. AMP-activated protein kinase: also regulated by ADP? Trends Biochem. Sci. 36: 470–477 [DOI] [PubMed] [Google Scholar]
- Hiltunen J. K., Mursula A. M., Rottensteiner H., Wierenga R. K., Kastaniotis A. J., et al. , 2003. The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 27: 35–64 [DOI] [PubMed] [Google Scholar]
- Hong S. P., Carlson M., 2007. Regulation of snf1 protein kinase in response to environmental stress. J. Biol. Chem. 282: 16838–16845 [DOI] [PubMed] [Google Scholar]
- Hong S. P., Leiper F. C., Woods A., Carling D., Carlson M., 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100: 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. P., Momcilovic M., Carlson M., 2005. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase alpha as Snf1-activating kinases in yeast. J. Biol. Chem. 280: 21804–21809 [DOI] [PubMed] [Google Scholar]
- Honigberg S. M., Purnapatre K., 2003. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116: 2137–2147 [DOI] [PubMed] [Google Scholar]
- Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., et al. , 2009. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23: 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A., French S. L., Tekotte H., Yerlikaya S., Stahl M., et al. , 2011. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 30: 3052–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey E. L., Shamji A. F., Bernstein B. E., Schreiber S. L., 2004. Rpd3p relocation mediates a transcriptional response to rapamycin in yeast. Chem. Biol. 11: 295–299 [DOI] [PubMed] [Google Scholar]
- Humston E. M., Dombek K. M., Tu B. P., Young E. T., Synovec R. E., 2011. Toward a global analysis of metabolites in regulatory mutants of yeast. Anal. Bioanal. Chem. 401: 2387–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E., Guo B., Arndt K. T., Schmelzle T., Hall M. N., 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Jambhekar A., Amon A., 2008. Control of meiosis by respiration. Curr. Biol. 18: 969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Medintz I., Michels C. A., 1997. Two glucose sensing/signaling pathways stimulate glucose-induced inactivation of maltose permease in Saccharomyces. Mol. Biol. Cell 8: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Carlson M., 1996. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 10: 3105–3115 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Broach J. R., 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18: 2782–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Davis C., Broach J. R., 1998. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 17: 6942–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Carlson M., 1992. Carbon regulation in Saccharomyces, in Molecular and Cellular Biology of the Yeast Saccharomyces, edited by J. R. Broach, J. R. Pringle, and E. W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Johnston M., Kim J. H., 2005. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans. 33: 247–252 [DOI] [PubMed] [Google Scholar]
- Johnston M., Hillier L., Riles L., Albermann K., Andre B., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XII. Nature 387: 87–90 [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Nishikawa J. L., Breitkreutz B. J., Tyers M., 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395–400 [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J. R., Schneper L., Broach J. R., et al. , 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18: 2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Yoshino K., Kondo C., Kawamata T., Oshiro N., et al. , 2010. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 30: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniak A., Xue Z., Macool D., Kim J. H., Johnston M., 2004. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 3: 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhumaa K., Wu B., Kielland-Brandt M. C., 2010. Conditions with high intracellular glucose inhibit sensing through glucose sensor Snf3 in Saccharomyces cerevisiae. J. Cell. Biochem. 110: 920–925 [DOI] [PubMed] [Google Scholar]
- Kensler T. W., Wakabayashi N., Biswal S., 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 47: 89–116 [DOI] [PubMed] [Google Scholar]
- Keys D. A., Lee B. S., Dodd J. A., Nguyen T. T., Vu L., et al. , 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10: 887–903 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Brachet V., Moriya H., Johnston M., 2006. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot. Cell 5: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevecz R. R., Bolen J., Forrest G., Murray D. B., 2004. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc. Natl. Acad. Sci. USA 101: 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosinska M. M., Crutchfield C. A., Bradley P. H., Rabinowitz J. D., Broach J. R., 2011. Yeast cells can access distinct quiescent states. Genes Dev. 25: 336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman L., Lemaire K., Ma P., Teunissen A. W., Donaton M. C., et al. , 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Kresnowati M. T., van Winden W. A., Almering M. J., ten Pierick A., Ras C., et al. , 2006. When transcriptome meets metabolome: fast cellular responses of yeast to sudden relief of glucose limitation. Mol. Syst. Biol. 2: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R., Muhe Y., Prick T., Bredschneider M., Bremer S., et al. , 2009. Piecemeal microautophagy of the nucleus: genetic and morphological traits. Autophagy 5: 270–272 [DOI] [PubMed] [Google Scholar]
- Kubler E., Mosch H. U., Rupp S., Lisanti M. P., 1997. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272: 20321–20323 [DOI] [PubMed] [Google Scholar]
- Kuepfer L., Peter M., Sauer U., Stelling J., 2007. Ensemble modeling for analysis of cell signaling dynamics. Nat. Biotechnol. 25: 1001–1006 [DOI] [PubMed] [Google Scholar]
- Kupiec M., Byers B., Esposito R. E., Mitchell A. P., 1997. Meiosis and sporulation in Saccharomyces cerevisiae, pp. 889–1036 in The Molecular and Cellular Biology of the Yeast Saccharomyces, edited by J. R. Pringle, J. R. Broach, and E. W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Kuttykrishnan S., Sabina J., Langton L. L., Johnston M., Brent M. R., 2010. A quantitative model of glucose signaling in yeast reveals an incoherent feed forward loop leading to a specific, transient pulse of transcription. Proc. Natl. Acad. Sci. USA 107: 16743–16748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferte A., Favry E., Sentenac A., Riva M., Carles C., et al. , 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 20: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan J., Mosley A. L., Ozcan S., 2003. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr. Genet. 44: 19–25 [DOI] [PubMed] [Google Scholar]
- Lamb T. M., Mitchell A. P., 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascaris R., Bussemaker H. J., Boorsma A., Piper M., van der Spek H., et al. , 2003. Hap4p overexpression in glucose-grown Saccharomyces cerevisiae induces cells to enter a novel metabolic state. Genome Biol. 4: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H., Whiteway M., 2008. Increased respiration in the sch9Delta mutant is required for increasing chronological life span but not replicative life span. Eukaryot. Cell 7: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. K., Shibata Y., Rao B., Strahl B. D., Lieb J. D., 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lee J., Moir R. D., Willis I. M., 2009. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J. Biol. Chem. 284: 12604–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P., Cho B. R., Joo H. S., Hahn J. S., 2008. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 70: 882–895 [DOI] [PubMed] [Google Scholar]
- Lee P., Paik S. M., Shin C. S., Huh W. K., Hahn J. S., 2011. Regulation of yeast Yak1 kinase by PKA and autophosphorylation-dependent 14–3-3 binding. Mol. Microbiol. 79: 633–646 [DOI] [PubMed] [Google Scholar]
- Leech A., Nath N., McCartney R. R., Schmidt M. C., 2003. Isolation of mutations in the catalytic domain of the snf1 kinase that render its activity independent of the snf4 subunit. Eukaryot. Cell 2: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire K., Van de Velde S., Van Dijck P., Thevelein J. M., 2004. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16: 293–299 [DOI] [PubMed] [Google Scholar]
- Lempiainen H., Uotila A., Urban J., Dohnal I., Ammerer G., et al. , 2009. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell 33: 704–716 [DOI] [PubMed] [Google Scholar]
- Lenssen E., James N., Pedruzzi I., Dubouloz F., Cameroni E., et al. , 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation–via a newly identified Glc7/Bud14 type I protein phosphatase module–and TFIID promoter distribution. Mol. Cell. Biol. 25: 488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J., 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6: 463–477 [DOI] [PubMed] [Google Scholar]
- Li H., Tsang C. K., Watkins M., Bertram P. G., Zheng X. F., 2006. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442: 1058–1061 [DOI] [PubMed] [Google Scholar]
- Liko D., Conway M. K., Grunwald D. S., Heideman W., 2010. Stb3 plays a role in the glucose-induced transition from quiescence to growth in Saccharomycescerevisiae. Genetics 185: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman S. I., Broach J. R., 2009. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl. Acad. Sci. USA 106: 19928–19933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu X., Kuo M. H., 2010. Snf1p regulates Gcn5p transcriptional activity by antagonizing Spt3p. Genetics 184: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Butow R. A., 1999. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19: 6720–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Butow R. A., 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40: 159–185 [DOI] [PubMed] [Google Scholar]
- Ljungdahl P. O., Daignan-Fornier B., 2012. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190: 885–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. S., Duggan L., Emre N. C., Belotserkovskya R., Lane W. S., et al. , 2001. Snf1–a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293: 1142–1146 [DOI] [PubMed] [Google Scholar]
- Lo W. S., Gamache E. R., Henry K. W., Yang D., Pillus L., et al. , 2005. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 24: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A., Hirsch J. P., 2005. Cyclic AMP-independent regulation of protein kinase A substrate phosphorylation by Kelch repeat proteins. Eukaryot. Cell 4: 1794–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Brauer M. J., Botstein D., 2009. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol. Biol. Cell 20: 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. Y., Lin Y. Y., Sheu J. C., Wu J. T., Lee F. J., et al. , 2011. Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell 146: 969–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke M. M., Della Seta F., Di Como C. J., Sugimoto H., Kobayashi R., et al. , 1996. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16: 2744–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P., Wera S., Van Dijck P., Thevelein J. M., 1999. The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol. Biol. Cell 10: 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B., Kaiser C. A., 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290: 1–18 [DOI] [PubMed] [Google Scholar]
- Malcher M., Schladebeck S., Mosch H. U., 2011. The Yak1 protein kinase lies at the center of a regulatory cascade affecting adhesive growth and stress resistance in Saccharomyces cerevisiae. Genetics 187: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion R. M., Regev A., Segal E., Barash Y., Koller D., et al. , 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101: 14315–14322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. E., Soulard A., Hall M. N., 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979 [DOI] [PubMed] [Google Scholar]
- Mayer C., Zhao J., Yuan X., Grummt I., 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer F. V., Heath R., Underwood E., Sanders M. J., Carmena D., et al. , 2011. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 14: 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney R. R., Schmidt M. C., 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276: 36460–36466 [DOI] [PubMed] [Google Scholar]
- Merz K., Hondele M., Goetze H., Gmelch K., Stoeckl U., et al. , 2008. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 22: 1190–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchelhill K. I., Stapleton D., Gao G., House C., Michell B., et al. , 1994. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J. Biol. Chem. 269: 2361–2364 [PubMed] [Google Scholar]
- Mok J., Kim P. M., Lam H. Y., Piccirillo S., Zhou X., et al. , 2010. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3: ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momcilovic M., Hong S. P., Carlson M., 2006. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J. Biol. Chem. 281: 25336–25343 [DOI] [PubMed] [Google Scholar]
- Momcilovic M., Iram S. H., Liu Y., Carlson M., 2008. Roles of the glycogen-binding domain and Snf4 in glucose inhibition of SNF1 protein kinase. J. Biol. Chem. 283: 19521–19529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H., Johnston M., 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 101: 1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H., Shimizu-Yoshida Y., Omori A., Iwashita S., Katoh M., et al. , 2001. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev. 15: 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16: 51–53 [DOI] [PubMed] [Google Scholar]
- Mosley A. L., Lakshmanan J., Aryal B. K., Ozcan S., 2003. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem. 278: 10322–10327 [DOI] [PubMed] [Google Scholar]
- Nachman I., Regev A., Ramanathan S., 2007. Dissecting timing variability in yeast meiosis. Cell 131: 544–556 [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R., Levy M., Tsechansky M., Stovall G. M., O’Connell J. D., et al. , 2009. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA 106: 10147–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath N., McCartney R. R., Schmidt M. C., 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23: 3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa T. K., Davis R. W., 2009. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 5: e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J., Cameron S., Toda T., Ferguson K. M., Wigler M., 1987. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1: 931–937 [DOI] [PubMed] [Google Scholar]
- Niranjan T., Guo X., Victor J., Lu A., Hirsch J. P., 2007. Kelch repeat protein interacts with the yeast Galpha subunit Gpa2p at a site that couples receptor binding to guanine nucleotide exchange. J. Biol. Chem. 282: 24231–24238 [DOI] [PubMed] [Google Scholar]
- Octavio L. M., Gedeon K., Maheshri N., 2009. Epigenetic and conventional regulation is distributed among activators of FLO11 allowing tuning of population-level heterogeneity in its expression. PLoS Genet. 5: e1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oficjalska-Pham D., Harismendy O., Smagowicz W. J., Gonzalez de Peredo A., Boguta M., et al. , 2006. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol. Cell 22: 623–632 [DOI] [PubMed] [Google Scholar]
- Orlova M., Kanter E., Krakovich D., Kuchin S., 2006. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot. Cell 5: 1831–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M., Ozcetin H., Barrett L., Kuchin S., 2010. Roles of the Snf1-activating kinases during nitrogen limitation and pseudohyphal differentiation in Saccharomyces cerevisiae. Eukaryot. Cell 9: 208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek S. P., Parikh A. S., Kron S. J., 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148: 893–907 [DOI] [PubMed] [Google Scholar]
- Palomino A., Herrero P., Moreno F., 2006. Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res. 34: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U., 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Parua P. K., Ratnakumar S., Braun K. A., Dombek K. M., Arms E., et al. , 2010. 14–3-3 (Bmh) proteins inhibit transcription activation by Adr1 through direct binding to its regulatory domain. Mol. Cell. Biol. 30: 5273–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I., Dubouloz F., Cameroni E., Wanke V., Roosen J., et al. , 2003. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 12: 1607–1613 [DOI] [PubMed] [Google Scholar]
- Peeters T., Louwet W., Gelade R., Nauwelaers D., Thevelein J. M., et al. , 2006. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. USA 103: 13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters T., Versele M., Thevelein J. M., 2007. Directly from Galpha to protein kinase A: the kelch repeat protein bypass of adenylate cyclase. Trends Biochem. Sci. 32: 547–554 [DOI] [PubMed] [Google Scholar]
- Pelaez R., Herrero P., Moreno F., 2010. Functional domains of yeast hexokinase 2. Biochem. J. 432: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T., Schuster S., Bonhoeffer S., 2001. Cooperation and competition in the evolution of ATP-producing pathways. Science 292: 504–507 [DOI] [PubMed] [Google Scholar]
- Philippi A., Steinbauer R., Reiter A., Fath S., Leger-Silvestre I., et al. , 2010. TOR-dependent reduction in the expression level of Rrn3p lowers the activity of the yeast RNA Pol I machinery, but does not account for the strong inhibition of rRNA production. Nucleic Acids Res. 38: 5315–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarska I., Rytka J., Rempola B., 2010. Regulation of sporulation in the yeast Saccharomyces cerevisiae. Acta Biochim. Pol. 57: 241–250 [PubMed] [Google Scholar]
- Pnueli L., Edry I., Cohen M., Kassir Y., 2004. Glucose and nitrogen regulate the switch from histone deacetylation to acetylation for expression of early meiosis-specific genes in budding yeast. Mol. Cell. Biol. 24: 5197–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela P., Howell S., Moreno S., Rossi S., 2002. In vivo and in vitro phosphorylation of two isoforms of yeast pyruvate kinase by protein kinase A. J. Biol. Chem. 277: 30477–30487 [DOI] [PubMed] [Google Scholar]
- Portela P., Moreno S., Rossi S., 2006. Characterization of yeast pyruvate kinase 1 as a protein kinase A substrate, and specificity of the phosphorylation site sequence in the whole protein. Biochem. J. 396: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers R. W., III, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S., 2006. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20: 174–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt Z. L., Drehman B. J., Miller M. E., Johnston S. D., 2007. Mutual interdependence of MSI1 (CAC3) and YAK1 in Saccharomyces cerevisiae. J. Mol. Biol. 368: 30–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S., Avila-Campillo I., Aldridge C., Srinivasan A., Dimitrov K., et al. , 2004. Control of yeast filamentous-form growth by modules in an integrated molecular network. Genome Res. 14: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., et al. , 2005. Global analysis of protein phosphorylation in yeast. Nature 438: 679–684 [DOI] [PubMed] [Google Scholar]
- Purnapatre K., Gray M., Piccirillo S., Honigberg S. M., 2005. Glucose inhibits meiotic DNA replication through SCFGrr1p-dependent destruction of Ime2p kinase. Mol. Cell. Biol. 25: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V., Shah K. H., Herman P. K., 2011. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell 43: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar S., Young E. T., 2010. Snf1 dependence of peroxisomal gene expression is mediated by Adr1. J. Biol. Chem. 285: 10703–10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar S., Kacherovsky N., Arms E., Young E. T., 2009. Snf1 controls the activity of adr1 through dephosphorylation of Ser230. Genetics 182: 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J. L., Iyer V. R., Brown P. O., Struhl K., 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6: 1297–1307 [DOI] [PubMed] [Google Scholar]
- Roberts D. N., Wilson B., Huff J. T., Stewart A. J., Cairns B. R., 2006. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell 22: 633–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., et al. , 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L. S., Fink G. R., 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95: 13783–13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J. R., Cardenas M. E., 2003. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S., Kumme J., Schuller H. J., 2004. Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr. Genet. 45: 121–128 [DOI] [PubMed] [Google Scholar]
- Rubenstein E. M., McCartney R. R., Zhang C., Shokat K. M., Shirra M. K., et al. , 2008. Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem. 283: 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin-Bejerano I., Sagee S., Friedman O., Pnueli L., Kassir Y., 2004. The in vivo activity of Ime1, the key transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae, is inhibited by the cyclic AMP/protein kinase A signal pathway through the glycogen synthase kinase 3-beta homolog Rim11. Mol. Cell. Biol. 24: 6967–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D., Zhao Y., Warner J. R., 2005. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 24: 533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabina J., Johnston M., 2009. Asymmetric signal transduction through paralogs that comprise a genetic switch for sugar sensing in Saccharomyces cerevisiae. J. Biol. Chem. 284: 29635–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I., Pinson B., Salin B., Daignan-Fornier B., 2006. Actin bodies in yeast quiescent cells: an immediately available actin reserve? Mol. Biol. Cell 17: 4645–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A. J., Brauer M. J., Botstein D., 2004. Nutritional homeostasis in batch and steady-state culture of yeast. Mol. Biol. Cell 15: 4089–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., et al. , 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier J. J., French S., Osheim Y., Cheung W. L., Gallo C. M., et al. , 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21: 4959–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo G. M., 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam A., Hartley A., Duvel K., Broach J. R., Garrett S., 2004. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot. Cell 3: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P., 2003. Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem. Soc. Trans. 31: 178–181 [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Ali S. M., Sabatini D. M., 2005. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17: 596–603 [DOI] [PubMed] [Google Scholar]
- Schawalder S. B., Kabani M., Howald I., Choudhury U., Werner M., et al. , 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432: 1058–1061 [DOI] [PubMed] [Google Scholar]
- Scherens B., Feller A., Vierendeels F., Messenguy F., Dubois E., 2006. Identification of direct and indirect targets of the Gln3 and Gat1 activators by transcriptional profiling in response to nitrogen availability in the short and long term. FEMS Yeast Res. 6: 777–791 [DOI] [PubMed] [Google Scholar]
- Schmidt M. C., McCartney R. R., 2000. beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 19: 4936–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. C., McCartney R. R., Zhang X., Tillman T. S., Solimeo H., et al. , 1999. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4561–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller H. J., 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43: 139–160 [DOI] [PubMed] [Google Scholar]
- Shamji A. F., Kuruvilla F. G., Schreiber S. L., 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10: 1574–1581 [DOI] [PubMed] [Google Scholar]
- Shirra M. K., McCartney R. R., Zhang C., Shokat K. M., Schmidt M. C., et al. , 2008. A chemical genomics study identifies Snf1 as a repressor of GCN4 translation. J. Biol. Chem. 283: 35889–35898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. J., Petti A. A., Slavov N., Parsons L., Briehof R., et al. , 2010. Metabolic cycling in single yeast cells from unsynchronized steady-state populations limited on glucose or phosphate. Proc. Natl. Acad. Sci. USA 107: 6946–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Tyers M., 2009. A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis. Genes Dev. 23: 1944–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N., Botstein D., 2011. Coupling among growth rate response, metabolic cycle, and cell division cycle in yeast. Mol. Biol. Cell 22: 1997–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smets B., Ghillebert R., De Snijder P., Binda M., Swinnen E., et al. , 2010. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 56: 1–32 [DOI] [PubMed] [Google Scholar]
- Smith A., Ward M. P., Garrett S., 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17: 3556–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielewoy N., Flick K., Kalashnikova T. I., Walker J. R., Wittenberg C., 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24: 8994–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland C. M., Hawley S. A., McCartney R. R., Leech A., Stark M. J., et al. , 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13: 1299–1305 [DOI] [PubMed] [Google Scholar]
- Swinnen E., Wanke V., Roosen J., Smets B., Dubouloz F., et al. , 2006. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C., Yoo J. Y., Tagne J. B., Kacherovsky N., Lee T. I., et al. , 2005. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol. Cell. Biol. 25: 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagkopoulos I., Liu Y. C., Tavazoie S., 2008. Predictive behavior within microbial genetic networks. Science 320: 1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Cooper T. G., 2003. Tor1/2 regulation of retrograde gene expression in Saccharomyces cerevisiae derives indirectly as a consequence of alterations in ammonia metabolism. J. Biol. Chem. 278: 36924–36933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Georis I., Feller A., Dubois E., Cooper T. G., 2009. Rapamycin-induced Gln3 dephosphorylation is insufficient for nuclear localization: Sit4 and PP2A phosphatases are regulated and function differently. J. Biol. Chem. 284: 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. J., Georis I., Dubois E., Cooper T. G., 2010. Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae. J. Biol. Chem. 285: 17880–17895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. M., Gaucher E. A., Burgan M. F., De Kee D. W., Li T., et al. , 2005. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat. Genet. 37: 630–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley R., Shapiro L., 2007. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science 315: 1726–1729 [DOI] [PubMed] [Google Scholar]
- Towpik J., Graczyk D., Gajda A., Lefebvre O., Boguta M., 2008. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J. Biol. Chem. 283: 17168–17174 [DOI] [PubMed] [Google Scholar]
- Tu B. P., Kudlicki A., Rowicka M., McKnight S. L., 2005. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science 310: 1152–1158 [DOI] [PubMed] [Google Scholar]
- Tu J., Carlson M., 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14: 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B., Liang X. B., Robert F., Soontorngun N., 2010. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 10: 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., et al. , 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26: 663–674 [DOI] [PubMed] [Google Scholar]
- Usaite R., Jewett M. C., Oliveira A. P., Yates J. R., 3rd, Olsson L., et al. , 2009. Reconstruction of the yeast Snf1 kinase regulatory network reveals its role as a global energy regulator. Mol. Syst. Biol. 5: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttenweiler A., Mayer A., 2008. Microautophagy in the yeast Saccharomyces cerevisiae. Methods Mol. Biol. 445: 245–259 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C., Thompson C. B., 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oevelen C. J., van Teeffelen H. A., van Werven F. J., Timmers H. T., 2006. Snf1p-dependent Spt-Ada-Gcn5-acetyltransferase (SAGA) recruitment and chromatin remodeling activities on the HXT2 and HXT4 promoters. J. Biol. Chem. 281: 4523–4531 [DOI] [PubMed] [Google Scholar]
- Vincent O., Carlson M., 1999. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 18: 6672–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O., Townley R., Kuchin S., Carlson M., 2001. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 15: 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Plehwe U., Berndt U., Conz C., Chiabudini M., Fitzke E., et al. , 2009. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev. 23: 2102–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J. T., Hall D. B., Struhl K., 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432: 1054–1058 [DOI] [PubMed] [Google Scholar]
- Wang G., Deschenes R. J., 2006. Plasma membrane localization of Ras requires class C Vps proteins and functional mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 3243–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Pierce M., Schneper L., Guldal C. G., Zhang X., et al. , 2004. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2: E128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson T., Schwartz J. M., Kell D. B., Stateva L., 2009. Deterministic mathematical models of the cAMP pathway in Saccharomyces cerevisiae. BMC Syst. Biol. 3: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I. M., Moir R. D., 2007. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem. Sci. 32: 51–53 [DOI] [PubMed] [Google Scholar]
- Wilson W. A., Hawley S. A., Hardie D. G., 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6: 1426–1434 [DOI] [PubMed] [Google Scholar]
- Wittner M., Hamperl S., Stockl U., Seufert W., Tschochner H., et al. , 2011. Establishment and maintenance of alternative chromatin states at a multicopy gene locus. Cell 145: 543–554 [DOI] [PubMed] [Google Scholar]
- Woods A., Munday M. R., Scott J., Yang X., Carlson M., et al. , 1994. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 269: 19509–19515 [PubMed] [Google Scholar]
- Wu B., Ottow K., Poulsen P., Gaber R. F., Albers E., et al. , 2006. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J. Cell Biol. 173: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuster A., Babu M. M., 2010. Transcriptional control of the quorum sensing response in yeast. Mol. Biosyst. 6: 134–141 [DOI] [PubMed] [Google Scholar]
- Xue Y., Batlle M., Hirsch J. P., 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 17: 1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Shen X., Jiang Y., 2006. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 25: 3546–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Lai Y., Jiang Y., 2012. The TOR complex 1 is a direct target of Rho1 GTPase. Mol. Cell 45: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Klionsky D. J., 2009. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335: 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Zaman S., Broach J. R., Klionsky D. J., 2007. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 18: 4180–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. T., Dombek K. M., Tachibana C., Ideker T., 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278: 26146–26158 [DOI] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Zhao X., Broach J. R., 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42: 27–81 [DOI] [PubMed] [Google Scholar]
- Zaman S., Lippman S. I., Schneper L., Slonim N., Broach J. R., 2009. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 5: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller C. E., Parnell S. C., Dohlman H. G., 2007. The RACK1 ortholog Asc1 functions as a G-protein beta subunit coupled to glucose responsiveness in yeast. J. Biol. Chem. 282: 25168–25176 [DOI] [PubMed] [Google Scholar]
- Zhao H., Wang J. Q., Shimohata T., Sun G., Yenari M. A., et al. , 2007. Conditions of protection by hypothermia and effects on apoptotic pathways in a rat model of permanent middle cerebral artery occlusion. J. Neurosurg. 107: 636–641 [DOI] [PubMed] [Google Scholar]
- Zhao Y., McIntosh K. B., Rudra D., Schawalder S., Shore D., et al. , 2006. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell. Biol. 26: 4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Byers K. J., McCord R. P., Shi Z., Berger M. F., et al. , 2009. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]