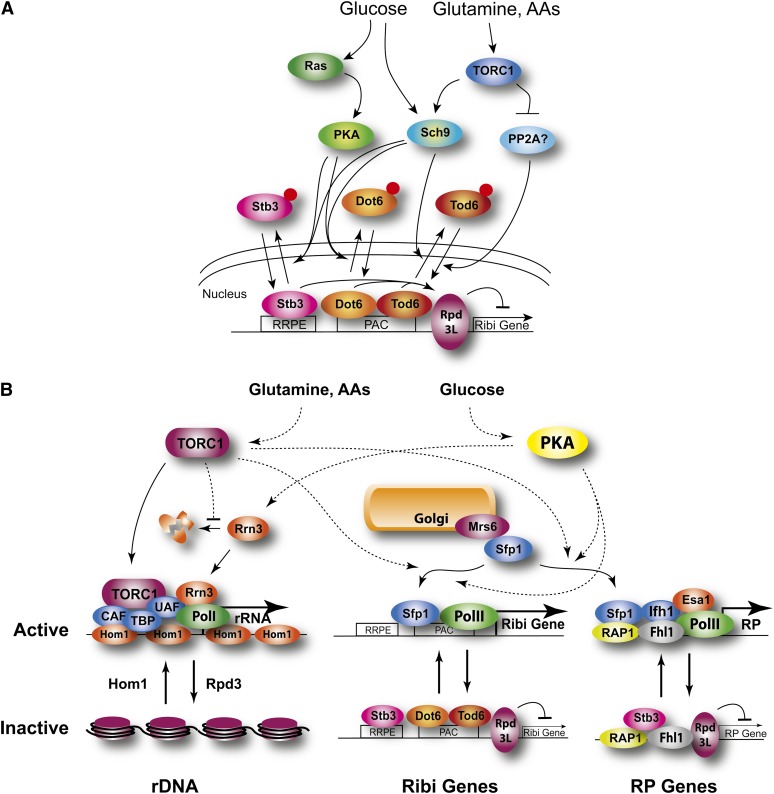
Figure 5 .
Regulation of ribosome biogenesis. (A) Ribi gene repressors. Ribi gene expression responds to nutritional input through alleviation of repression effected by Dot1, Tod1, and Stb3, which recruit the histone deacetylase Rpd3L. Expression requires inactivation of all three repressors and, while input exhibits significant cross-talk, glucose and Ras/PKA predominantly influence Dot1 activity while nitrogen and TORC1 predominantly influence Tod6 activity. (B) rDNA, RP, and Ribi gene regulation. Regulation of rRNA transcription by nutrients involves both template activation—from a repressed, nucleosome-bound state to a locus bound predominantly by the high-mobility group protein Hmo1—as well as regulation of Pol I initiation, primarily controlled by the level and interaction of Rrn3 with Pol I. Transcriptional regulation of Ribi and ribosomal protein (RP) genes involves both local reorganization of the promoters as well as translocation of the split finger transcription factor from a cytoplasmic association with Mrs6 to the nucleus, where it peripherally associates with the promoters.