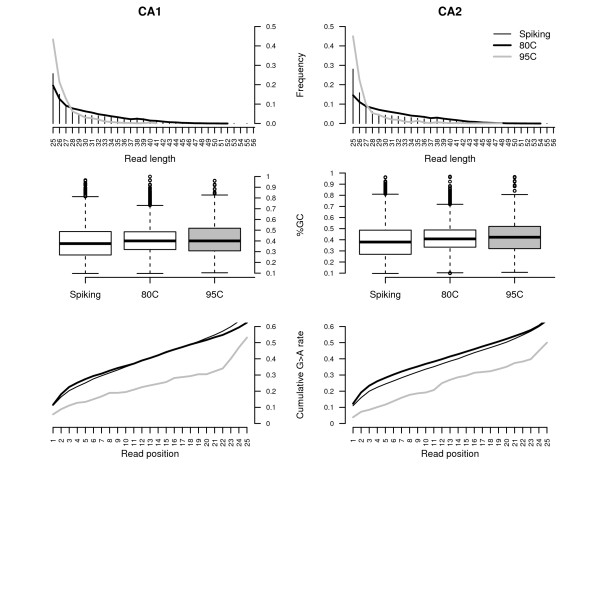
Figure 1.
Sequence features of the population of tSMS reads recovered from sample CA (>50,300 BP). Sample CA was extracted in duplicate and both extracts were tSMS sequenced on a Helicos 1100 FOV channel following different template preparation procedures as described in the methods section. Top: Read length distribution (spiked sequences are reported as vertical bars). Middle: Read %GC contents. Right: Cumulative guanine to adenine misincorporation rates as a function of the distance from sequencing start. This class of mismatch derives from the post-mortem deamination of cytosine residues and can be taken as a proxy for post-mortem DNA damage.