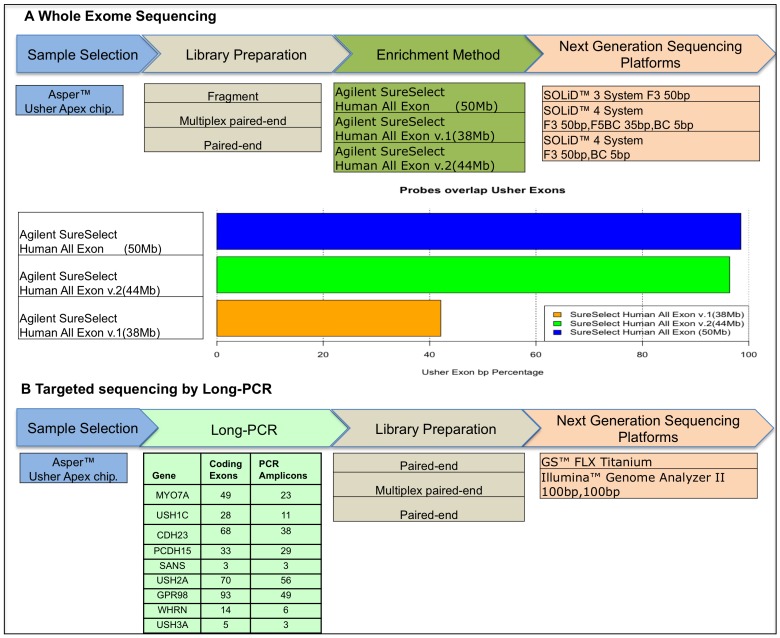
Figure 1. Workflow of the next generation sequencing strategies used.
A) whole exome sequencing workflow. Samples have been pre-screened using an Apex-based Usher genotyping microarray; library preparations prior to enrichment include fragment single reads or Paired-End preparation. Three different types of enrichment methods have been used; each enrichment probe sets overlap at different extent to the RefSeq coding regions of Usher genes (horizontal bars). Sequencing protocols include single 50 bp reads on the Solid3 System, single 50 bp read on Solid4 System, Paired-end reads 50 bp+35 bp on Solid4 System. B) Long-PCR sequencing workflow. Samples have been pre-screened using Usher Apex microarray, Long-PCR approach produced 218 PCR amplicons used as input for the for Fragment and Paired-End library preparation. Sequencing was performed using both GS-FLX and GAII Systems.