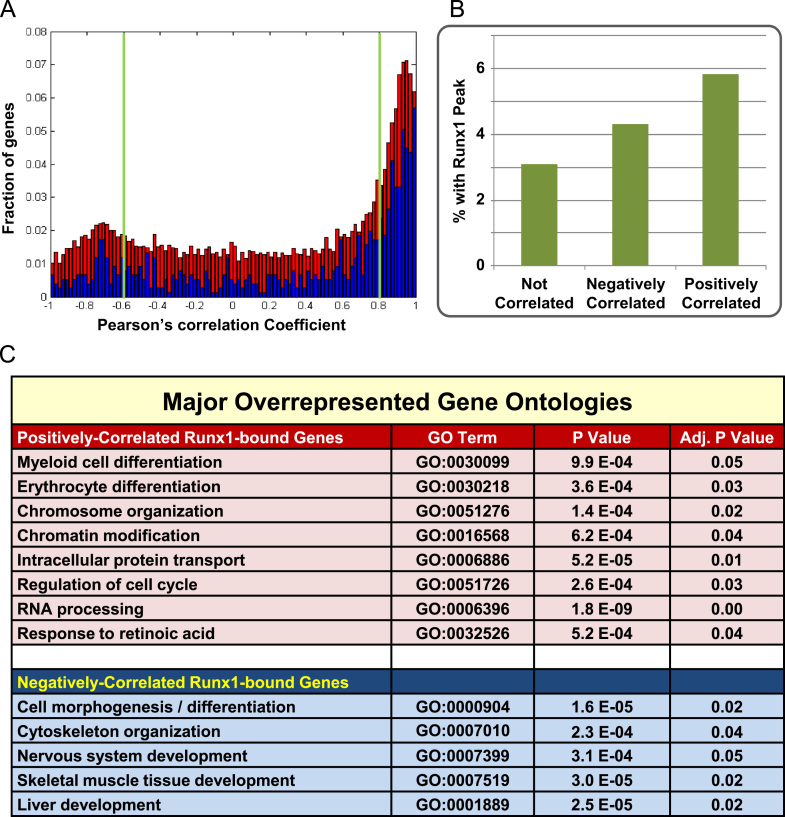
Fig. 5.
Integration of expression and ChIP-Seq datasets defines a candidate Runx1 target gene set. (A) Histogram showing correlation with Runx1 dynamics for all genes (red) and Runx1 bound genes (blue) (scaled appropriately for superposition) with correlation (0.8) and negative correlation (−0.6) cut-offs. (B) Fraction of Runx1 bound genes in not-correlated, negatively correlated and correlated gene- sets respectively (as defined using the cut-offs in (A). The gradual increase supports the regulatory role of Runx1 inferred from binding events. (C) Analysis of putative gene functions using gene ontology overrepresentation (Al-Shahrour et al., 2008) reveals biological functions consistent with haematopoietic development for correlated Runx1-bound genes, and non-haematopoietic fates for negatively correlated Runx1 bound genes. See Supplementary Fig. 3 for Gene Ontology analysis results using peak regions from single populations.