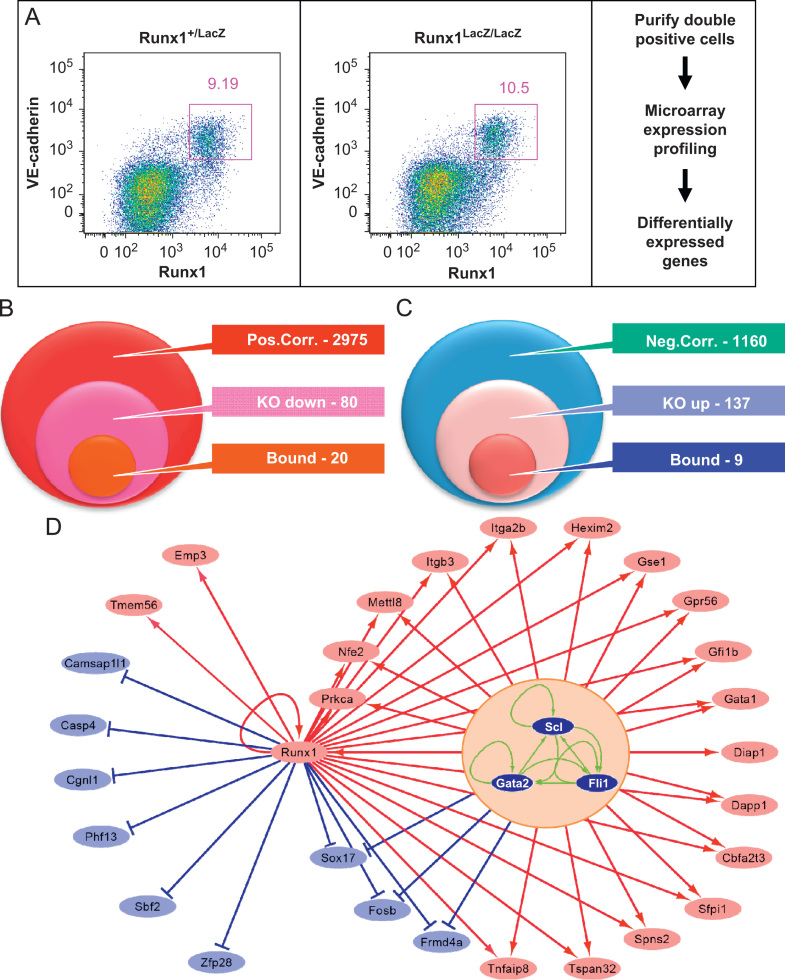
Fig. 6.
Intersection of Runx1-correlated and Runx1-bound genes with Runx1−/− expression profiling suggests a step-wise establishment of the haematopoietic transcriptional programme. (A) Identification of differentially expressed genes in E7.5 Runx1+/− and Runx1−/− embryos. VE-cadherin+/Runx1+ cells were flow sorted from E7.5 mouse embryos and differentially expressed genes were identified by microarray expression profiling. The pink square indicates the gate used for cell sorting. (B) Intersection of gene expression and ChIP-Seq datasets identifies a candidate gene set positively controlled by Runx1. ‘Pos.Corr.’, genes positively correlating with Runx1 expression from Fig. 5(A) (e.g., using expression profiling data from in vitro differentiated ES cells); ‘KO down’, genes at least 1.5-fold lower in E7.5 Runx1−/− than Runx1+/− from (A) (e.g., using primary cells derived from mouse embryos); ‘Bound’, genes bound by Runx1 in one or more of the three populations from Fig. 3C) Intersection of gene expression and ChIP-Seq datasets identifies a candidate gene set negatively controlled by Runx1. ‘Neg.Corr.’, genes negatively correlating with Runx1 expression from Fig. 5(A) (e.g., using expression profiling data from in vitro differentiated ES cells); ‘KO up’, genes at least 1.5-fold higher in E7.5 Runx1−/− than Runx1+/− from A (e.g., using primary cells derived from mouse embryos); ‘Bound’, genes bound by Runx1 in one or more of the 3 populations from Fig. 3D) The majority of Runx1 positively controlled genes from Fig. 6B are bound by the Scl/Gata2/Fli1 triad in haematopoietic progenitor cells. The diagram shows all Runx1-positively controlled genes in red and negatively controlled genes in blue. Links are also drawn for those genes bound by the Scl/Gata2/Fli1 triad in haematopoietic progenitor cells, showing binding for 18/20 Runx1 positively controlled and 3/10 Runx1-negatively controlled genes. Only one of the genes (Mettll3) has a Runx1 peak bound in populations 2–4.