Abstract
An Arabidopsis thaliana mutant, cbd (carotenoid biosynthesis deficient), was recovered from a mutant population based on its yellow cotyledons, yellow-first true leaves, and stunted growth. Seven-day-old seedlings and mature seeds of this mutant had lower chlorophyll and total carotenoids than the wild type (WT). Genetic and molecular characterization revealed that cbd was a recessive mutant caused by a T-DNA insertion in the gene cpSRP54 encoding the 54kDa subunit of the chloroplast signal recognition particle. Transcript levels of most of the main carotenoid biosynthetic genes in cbd were unchanged relative to WT, but expression increased in carotenoid and abscisic acid catabolic genes. The chloroplasts of cbd also had developmental defects that contributed to decreased carotenoid and chlorophyll contents. Transcription of AtGLK1 (Golden 2-like 1), AtGLK2, and GUN4 appeared to be disrupted in the cbd mutant suggesting that the plastid-to-nucleus retrograde signal may be affected, regulating the changes in chloroplast functional and developmental states and carotenoid content flux. Transformation of A. thaliana and Brassica napus with a gDNA encoding the Arabidopsis cpSRP54 showed the utility of this gene in enhancing levels of seed carotenoids without affecting growth or seed yield.
Key words: Accumulation, Arabidopsis thaliana, biosynthesis, Brassica napus, carotenoids, gene expression
Introduction
Carotenoids are essential components of the photosynthetic machinery and play a critical role in preventing photooxidative damage (Howitt and Pogson, 2006). Carotenoids are precursors of the plant hormone abscisic acid (ABA) (McCarty, 1995). They are also precursors of vitamin A, and some carotenoids protect against human age-related macular degeneration (Beatty et al., 2004). A few carotenoids are used as colourants in the food and cosmetics industries and some are important supplements in fish feed formulations (Umeno et al., 2005).
Carotenoids are derived from the isoprenoid pathway and are synthesized in almost all types of plant plastids (Howitt and Pogson, 2006). They co-accumulate with chlorophyll in pigment-binding protein complexes embedded in thylakoid membranes (Botella-Pavía and Rodríguez-Concepción, 2006). Generally, the amounts of β-carotene and lutein are proportional to the content of chlorophyll a and b, respectively (Pogson et al., 1996). Carotenoid-deficient mutants usually show impaired photosynthesis (reviewed by DellaPenna, 1999; Dong et al., 2007)
Three independent plastid-to-nucleus retrograde signalling pathways have been described (Nott et al., 2006). The best-studied retrograde pathway is triggered by the accumulation of the chlorophyll precursor, Mg-Protoporphyrin IX (Mg-ProtoIX), in the tretrapyrrole pathway (Larkin et al., 2003; Strand et al., 2003). GUN4 (GENOMES UNCOUPLED 4) is a chloroplast-encoded Mg-ProtoIX-binding protein and can stimulate Mg-chelatase activity (Larkin et al., 2003). GUN1 encodes a chloroplast-localized pentatricopeptide repeat protein, and the three known chloroplast retrograde signal pathways intersect upstream of GUN1 (Koussevitzky et al., 2007). AtGLK1 and AtGLK2 (Golden 2-Like) are redundant members of the GARP superfamily of transcription factors (Fitter et al., 2002). AtGLK1 is downregulated by GUN1 when plastids are dysfunctional and has been reported as a positive regulator of photosynthesis-related nuclear gene expression in a plastid-to-nucleus signalling pathway (Kakizaki et al., 2009). ABI4 acts as a negative regulator of photosynthesis-related nuclear gene expression and part of the GUN4/GUN1 signalling pathway (Koussevitzky et al., 2007).
Although carotenoid biosynthesis pathways in plants have been well documented (Howitt and Pogson, 2006), the mechanisms that regulate carotenoid biosynthesis and accumulation in plants are less understood. A combination of factors are known to regulate carotenoid accumulation in plants. These include metabolic controls that determine the rate of anabolism and catabolism, availability of storage compartments, and transcriptional and epigenetic regulation (Hannoufa and Hossain, 2012). To gain insight into additional mechanisms, an activation tagged population of Arabidopsis thaliana was screened, and this led to the identification of a new T-DNA insertion mutant named cbd (carotenoid biosynthesis deficiency). The phenotype of the cbd mutant was shown to be due to a mutation in the chloroplast signal recognition particle 54kDa subunit gene (cpSRP54). The mutation reduced chlorophyll and carotenoid content, impaired chloroplast development, and caused an ABA deficiency in seedlings of cbd. These data showed that over-expression of cpSRP54 in Arabidopsis and Brassica napus raised carotenoid levels in seeds without affecting growth or seed yield and suggested that cpSRP54 can be used as a tool for biofortification of carotenoids in oilseed crops and other crop plants.
Materials and methods
Arabidopsis plant materials and identification of carotenoid-deficient mutants
A. thaliana (ecotype Columbia) T-DNA activation tagged SK mutant population was generated using the pSKI015 vector at Agriculture and Agri-Food Canada (Robinson et al., 2009; www.brassica.ca). The cpSRP54 mutants, CS850421 and SALK_079668 (Alonso et al., 2003), were obtained from the Arabidopsis Biological Resource Center (ABRC, www.biosci.ohio-state.edu/~plantbio/Facilities/abrc/abrchome.htm). The mutants were analysed by PCR amplification using one primer specific to the T-DNA (LBb1, Table 1) and two primers specific to the flanking genomic DNA sequence (P1 and P2 for CS850421; P3 and P4 for SALK_079668). Homozygous Arabidopsis mutant seeds were used for further studies.
Table 1.
Primers used to characterize Arabidopsis cbd mutant
| Primers used for genotyping and molecular complementation | ||||
| Primer | AGI code | Sequence (from 5′ to 3′) | Purpose | |
| pSKTAIL-L1 | N/A | TTCTCATCTAAGCCCCCATTTGG | Confirmation of T-DNA insertion site in cbd mutant | |
| LBb1 | N/A | GCGTGGACCGCTTGCTGCA | Confirmation of T-DNA insertion site in SALK lines | |
| P1 | AT5G03940 | ACCGGTTCTTGTTTTTGTTTCATTAGCA | Confirmation of T-DNA insertion site in CS850421 | |
| P2 | AT5G03940 | CATTCGCTCTCCACGTCCTACCAGTTTA | Confirmation of T-DNA insertion site in CS850421 | |
| P3 | AT5G03940 | AGCCCCATTGTCAGGTTATTTCGCA | Confirmation of T-DNA insertion site in SALK_079668 | |
| P4 | AT5G03940 | GAGCTCACGGGCAACGAAATAAGAAAGA | Confirmation of T-DNA insertion site in SALK_079668 | |
| P5 | AT5G03940 | TATTATGGATACTGCAGGGAGGCTTCA | Confirmation of T-DNA insertion site in cbd | |
| P6 | AT5G03940 | TTGTTCTGCTTTCAATGCGTCCTCG | Confirmation of T-DNA insertion site in cbd | |
| P7 | AT5G03940 | ATGGAGGCTCTTCAATTTTCCAGCGT | Forward primer for amplification of cpSRP54 coding region | |
| P8 | AT5G03940 | GTTACCAGAGCCGAAGCCACGAGGA | Reverse primer for amplification of cpSRP54 coding region | |
| P9 | AT5G03940 | GTTTTCCTTTGGTTCAAATTTCCATTTA | Forward primer for amplification of cpSRP54 genomic sequence | |
| P10 | AT5G03940 | GTATTTTAGTTATGCAAAGCCTATGAA | Reverse primer for amplification of cpSRP54 genomic sequence | |
| AtACT2-F | AT3G18780 | GATATGGAAAAGATCTGGCATCAC | Forward primer for amplification of AtACT2 | |
| AtACT2-R | AT3G18780 | TCATACTCGGCCTTGGAGATCCAC | Reverse primer for amplification of AtACT2 | |
| Gene-specific primers used in quantitative real-time PCR | ||||
| Gene | AGI code | Forward primer (from 5′ to 3′) | Reverse primer (from 5′ to 3′) | Reference |
| PSY | AT5G17230 | TGCGGTGAAGTTTGCGCTGA | TGAAGCATTTGGCCCATCCA | |
| PDS | AT4G14210 | GTCGGTCACGCGCTCAGGTA | CGAGATGCTGACATGGCCAGA | |
| ZDS | AT3G04870 | CCATCGTCACGAGGCCTAGAA | TGTGTATGAACCGGCGAGGA | |
| bLYC | AT3G10230 | TGGTAGCGCTGCTCTTTTGGA | ACCAGCAGGACCACCACCAA | |
| BCH1 | AT4G25700 | GGCACGCTTCTCTATGGAATATGCATGA | GAATCCATAAGAGAGGAGACCAATCGCT | |
| BCH2 | AT5G52570 | ATGGAGTTTTGGGCAAGATGGGCTCAT | CAGGAACCGCGTTTGTTATAGCAAACACA | |
| LUT1 | AT3G53130 | CGAAATCCCAATCATGGGTCA | GCACCTCCGAGGAGATCAGC | |
| ZEP | AT5G67030 | ATGACCGGCTTCGAGAGTGG | TTCCGACGATGCAAGGTTGA | |
| VDE | AT1G08550 | ACCGCTCCGCTGTTGCTAAA | TGGCAATGCACTTTGCGAGT | |
| CCD1 | AT3G63520 | TGTTCCGCGTGAGACAGCAG | CCACCACTGCCACCGGTTC | |
| CCD4 | AT4G19170 | AAGATCTCCGGTGTGGTGAAGC | CCGGATTACCAGGATCCCTAGC | |
| CCD7 | AT2G44990 | CTAAACCGTGGCGACGACAA | CCGGAAAATCTGACGGCTTG | |
| CCD8 | AT4G32810 | CGTTTATGCATGCGGTGCTC | GGTCGAGGCACGAAGAATGG | |
| CAO | AT1G44446 | CCGGTGGAACACGGTTTACTTCTAGATA | AGTATCCTTGGAGACCCGAGGTAGGTGT | |
| PORA | At1g03630 | CTCGGTGTTTCAACCTTTGG | CAGATAGAATCAGCCAAAACACAAC | Kakizaki et al. (2009) |
| PORB | AT4G27440 | TTCCGAGAGCACATTCCTCTCTTCCGT | CCCTGATTTCGTCAAGCTTGGATCACT | |
| Lhca2 | AT3G61470 | ACATCTACACTGGCACTGGTCCTATTGA | ACACACAAACGCATTCACCTCCCCATAA | |
| Lhcb1 | AT1g29930 | AGCTCAAGAACGGAAGATTGG | GCCAAATGGTCAGCAAGGTT | Kakizaki et al. (2009) |
| Lhcb2.1 | AT2g05100 | AAGTCGTGAATGTACTTATTGGTG | GGTGGTGTGGTTCATTAAAGGT | Kakizaki et al. (2009) |
| LSU | ATCg00490 | TTGCCGAGATAATGGCCTACTT | AACAAAGCCCAAAGTTGACTCC | Kakizaki et al. (2009) |
| OE33 | AT5G66570 | ACCGTCAAGGCAGACAGTGTAAGCAAGA | CTTGAAATTGACGCTTCCGTCTGAAGCA | |
| PSBA | ATCG00020 | GTGTATTCGGCGGCTCCCTTTTTAGT | CTTCTTCTTGCCCGAATCTGTAACCTTCA | |
| PSBS | AT1G44575 | CATTGGAGCTCTCGGAGACAGAGGAA | CTCGTTCGCCTTCGTGAACCCAAACAAT | |
| NCED3 | AT3g14440 | TAACGCCGTTAGCTTAGAGGTTGAAGCA | AGAACTCACACGACCTGCTTCGCCAAAT | |
| NCED5 | AT1G30100 | TGTTCACGACGAGGAGAGTTGG | TATCCGCCGAATTCACGAAAGT | |
| NCED6 | AT3G24220 | GAGCTGGGATCGGTCTAGTGGA | TTGACCGTCGATCTTCACTTGG | |
| NCED9 | AT1G78390 | TCGACGGAGACGGTATGGTACA | TCTCCAATTGCTTTGGGGAAAA | |
| ABA2 | AT1G52340 | ACGGTTGATGATGTAGCGAACGCTGTT | CATCTGAAGACTTTAAAGGAGTGGTTAG | |
| AAO3 | AT2G27150 | TGGACTGCTCCTTCTGGTGATG | CTTGATGCTTCTTGGCGAGACA | |
| CYP707A1 | AT4G19230 | ATTTGATCCATCAAGATTCGAGGTGGCT | ACCAAACCTGTACTTGGTGGTGAGATGA | |
| CYP707A2 | AT2G29090 | GCGAATCCATCACTCCTCCGAATTCTT | TCACTTCCTGGACATGAGTGCACTCCAT | |
| CYP707A3 | AT5G45340 | CGAGATTCGAAGTTGCGCCGAAACCGA | GATTGACCATCTGTACTTAGTGGTGAGA | |
| CYP707A4 | AT3G19270 | GTGTGCTAACCCAAGAACAGATTGCAGA | CAGCCTTAACAGCTTCTAGAAGTTTCTGA | |
| At5g12240 | At5g12240 | TGGCACGATGCACCGACTGTTGCT | CTAGTCAATCTAACA AGCCAGTAAGCTA | Czechowski et al. (2005) |
| UBC | At5g25760 | TGCTTGGAGTCCTGCTTGGA | TGTGCCATTGAATTGAACCCTCT | Czechowski et al. (2005) |
SK mutants with defects in carotenoid accumulation were identified by sowing approximately 1000 seeds from each T2 super pool (37 pools, 100 lines per pool) on RediEarth soil (WR Grace, Ajax, Canada) and growing the lines in a controlled greenhouse environment (light intensity 230 µmol m–2 s–1; 16/8 light/dark; 20/17 °C). Arabidopsis seedlings with light green or pale yellowish leaves were selected as putative carotenoid mutants. For subsequent analysis, sterilized homozygous mutant seeds were incubated in 0.05% agarose at 4 °C for 2 days, then placed on Petri dishes containing 1/2 Murashige and Skoog basal medium (MS), 1% sucrose, and 0.8% agar and incubated in a growth cabinet at 60–80 µmol.m–2.s–1 under 16/8 light/dark at 22 °C. Seven-day-old Arabidopsis seedlings were used for carotenoid analysis, semi-quantitative reverse transcription PCR (RT-PCR), quantitative real-time PCR (qRT-PCR), and ABA quantification. For carotenoid analysis and qRT-PCR analysis of cpSRP54 gene expression in transgenic Arabidopsis plants, 8-day-old seedlings grown under the same growth cabinet conditions as above were used.
Molecular characterization of the Arabidopsis cbd mutant, cbd complementation lines, and wild-type over-expression lines
Genomic DNA was isolated using the method of Michiels et al. (2003), and used for thermal asymmetric interlaced (TAIL)-PCR (Liu et al., 1995) and genome walking (BD GenomeWalker Universal Kit, BD Biosciences Clontech, Mississauga, ON, Canada) to identify T-DNA insertion sites in the mutant line. The results were confirmed by PCR with one T-DNA-specific primer, pSKTAIL-L1, and two primers, P5 and P6, specific to the flanking genomic DNA sequence (Table 1).
For molecular complementation of mutant line cbd and development of cpSRP54 over-expression lines in a wild-type (WT) background, a 6.2-kb region of genomic DNA (gDNA) including the native promoter, 5′-untranscribed region (UTR), coding region, 3′-UTR, and 3′ flanking sequence of cpSRP54 (At5g03940), was amplified by PCR with primers P9 and P10 (Table 1), cloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen, Burlington, ON, Canada), and then subsequently subcloned into the binary vector pMDC99 (Curtis and Grossniklaus, 2003). Primers P7 and P8 (Table 1) were used to amplify the full-length cpSRP54 coding region (CDS), which was cloned in an identical cloning vector and subsequently subcloned into the binary vector pMDC83 (Curtis and Grossniklaus, 2003). Binary plasmids were introduced into Agrobacterium tumefaciens strain GV3101pMP90, and A. thaliana transformation was conducted according to the floral dip procedure (Clough and Bent, 1998).
RNA isolation, semi-quantitative RT-PCR, and qRT-PCR of Arabidopsis tissues
Total RNA was isolated from mutant, WT, and complemented/over-expression transgenic Arabidopsis plants using the RNeasy Plant Mini Kit (Qiagen, Mississauga, ON, Canada) and used for RT-PCR analysis of cpSRP54 gene expression and co-amplification of the internal control gene, AtACT2 (AT3G18780) (primers listed in Table 1). cDNA synthesis was conducted using 1 µg of total RNA and SuperScriptII reverse transcriptase (Invitrogen). PCR amplification was performed using an initial denaturation at 94 °C for 4min, followed by 26 cycles at 94 °C for 25 s, 55 °C for 30 s, and 72 °C for 1min 45 s, and a final extension at 72 °C for 7min. The amplicon was confirmed by sequencing.
For qRT-PCR, total RNA and cDNA preparations were made as described above. cDNA samples were diluted 10-fold and 2 µl diluted cDNA was used in each PCR reaction. qRT-PCR reactions were performed on three cDNA batches with a Bio-Rad CFX Manager System (Bio-Rad Laboratories, Mississauga, ON, Canada) using 12.5 µl of 2× SYBR Green qPCR Master Mix (Invitrogen) and gene-specific primers listed in Table 1. Amplifications of the ubiquitin-conjugating enzyme gene (UBC) and At5g12240 (Czechowski et al., 2005) were used as internal controls for normalization of expression levels. Amplification efficiency for internal controls and all target genes were 100%. Triplicate seedling samples were used and analysed statistically using ANOVA.
B. napus transformation and plant growth conditions
Arabidopsis cpSRP54 genomic sequence including 5′- and 3′-UTR was amplified by PCR using primers FFC-F31 and FFC-R34 (Table 2) with added BamHI and SalI sites, respectively, and TA-cloned into vector pCR2.1-TOPO for sequencing. Subsequently the cpSRP54 fragment was subcloned into the BamHI and SalI sites of the modified binary vector pBI121 in which the CaMV 35S promoter was replaced with the seed-specific B. napus napin promoter (Rask et al., 1998) at the BamHI and HindIII sites. Cotyledons with cotyledonary petioles from 5-day-old seedlings of B. napus doubled haploid line DH12075 were used as explants for transformation with A. tumefaciens GV3101 pMP90 harbouring the cpSRP54 + construct. One ml of the overnight A. tumfaciens culture was added to liquid MS media containing 1mg l–1 2,4-dichlorophenoxyacetic acid, 0.5g l–1 MES, 1% DMSO, and 0.04g l–1 acetosyringone buffered at a pH 5.6. B. napus explants were floated in this A. tumfaciens suspension for 2 hours after which the explants were removed, placed on filter paper moistened with MS media, incubated in the dark at 15 °C for 5 days, and transferred to selection media containing 0.7% Phytablend agar (Caisson Laboratories, North Logan, UT, USA), 3% sucrose, 4.5mg l–1 benzylaminopurine, 450mg l–1 timentin, and 20mg l–1 kanamycin. After 4–6 weeks, regenerated green shoots were transferred every 2 weeks to shoot selection media with BA at 0.5mg l–1 until normalized, then placed in 100ml rooting jars containing MS medium with 3% sucrose, 0.1mg l–1 NAA, 0.7% Phytablend agar, 450mg l–1 timentin, and buffered to pH 5.8. Rooted shoots were transferred to pots in a controlled environment greenhouse 16/8 light/dark at 20/17 °C. Only those plants with a confirmed transgene as determined by PCR using napin-F5 and FFC-R6 primers (Table 2) were subjected to further analysis.
Table 2.
Primers used in this study to generate and characterize transgenic cpSRP54 + B. napus lines
| Primers used for vector construction and confirmation of transgenic lines | |||
| Primer name | Sequence (from 5′ to 3′) | Purpose | |
| FFC-F31 | ggatccGTACCAAGAA GATTGCGGAG | Cloning of cpSRP54 (FFC) genomic sequence, add BamHI site (in lower case) | |
| FFC-R34 | gtcgacGTATTTTAGTTATGCAAAGCCTATGAA | Cloning of cpSRP54 (FFC) genomic sequence, add SalI site (in lower case) | |
| napin-F5 | AGCTCCCAATTTATATTCCCAACGGCAC | Confirmation of transgenic lines | |
| FFC-R6 | GAGCTCACGGGCAACGAAATAAGAAAGA | Confirmation of transgenic lines | |
| Gene-specific primers used in quantitative real-time PCR | |||
| Gene name | Forward primer (from 5′ to 3′) | Reverse primer (from 5′ to 3′) | Reference |
| cpSRP54 | ACAAAAGGCTCCACCTGGAACTGC | AGCCGAAGCCACGAGGACCA | Fujisawa et al. (2009) |
| BnbLYC | TCCACTGTTGTCTGCAGTGACG | CATCGACCTCAGCAACGATACC | Fujisawa et al. (2009) |
| BneLYC | TGAGGAGGTGTGTGGAGTCAGG | GAAGCTGCTCCAGAAGCAACAG | Fujisawa et al. (2009) |
| BnBCH | CAGAGGCTTCTCGGTCTGCTAC | CCTCTCGGACTTCTTCCTCTCC | Fujisawa et al. (2009) |
| BnCRTISO | GAGGTGGCAGCTGGAATCATAC | TCCTCTTGGCATTGGTCCATAC | Fujisawa et al. (2009) |
| BnPDS | GGCTGCAGTGGAAGGAACACTC | TCTCTGGCCATGTCAGCATCTC | Fujisawa et al. (2009) |
| BnPSY | CCAAAGCAACGACCGAGAGTG | CATCTGAGAGACCAGCCTGAGC | Fujisawa et al. (2009) |
| BnVDE | TCACGACCGTACGAGATTCTTC | AATCCAGATAAGGGTCGTGAGG | Fujisawa et al. (2009) |
| BnZDS | GCAATGAAAGACATTCGCAACC | TCTCGCACTCATGTTGTCACAG | Fujisawa et al. (2009) |
| BnZEP | TGCTGAAGAAGTCATGGAAGCTG | CTGCTAATCACCCGAGTCACAGG | Fujisawa et al. (2009) |
| BnACT3 | GCATCCCTCAGCACTTTCCAACAGA | ACCACGAACCAGAAGGCAGAAACACT | |
RNA extraction and qRT-PCR analysis of developing B. napus seeds
Total RNA was isolated from developing B. napus seeds at 34 days post anthesis (DPA) as described by Suzuki et al. (2004) with some modifications. Briefly, the seeds were ground into fine powder in liquid nitrogen. Approximately 50mg seeds were used for RNA extraction with 700 µl of extraction buffer. After extraction with chloroform/isoamylalcohol (CIA, 24:1) and a mixture of water-saturated phenol containing 35% (w/v) guanidium thiocyanate and 1/10 (v/v) 2M sodium acetate (pH 4.0), the supernatant was transferred to a new tube and a half volume of 100% ethanol was added to precipitate RNA. Subsequent procedures were performed according to the manufacturer’s instructions for the RNeasy Plant Mini Kit (Qiagen). cDNA synthesis and qRT-PCR reactions were conducted as described for Arabidopsis tissue except that 1 µl of 10-fold diluted cDNA was used in each PCR reaction. Amplification of B. napus BnACT3 was used as an internal control for the normalization of expression levels. For each independent transgenic line, triplicate seed samples were tested for expression and analysed statistically using ANOVA.
Extraction and analysis of carotenoids from seeds and seedlings
A. thaliana and B. napus seeds were harvested immediately upon maturation of soil-grown plants and stored at –20 °C. Approximately, 150mg seeds or 50mg agar-grown seedlings was used for carotenoid analysis. Seed carotenoid extraction and HPLC analysis were conducted as described previously (Yu et al., 2007). For carotenoid analysis of seedlings, saponification was conducted at room temperature for 30min. Procedures of extraction and HPLC analysis following saponification were the same for seedlings and seeds. Triplicate samples were used for carotenoid analysis and analysed statistically using ANOVA.
Chlorophyll extraction and quantification in Arabidopsis
Seven-day-old Arabidopsis seedlings (20–40mg freshweight) were ground into fine powder with liquid nitrogen. The sample powder was suspended in 1 ml of ice-cold 80% acetone and centrifuged at 13,000 g for 10min at 4 °C. The chlorophyll content of the supernatant was measured at 647nm and 664nm, and calculated using the formula described by Inskeep and Bloom (1985).
Detection of cell death and histochemical detection of O2 – in Arabidopsis
Arabidopsis WT and mutant seeds were sterilized and grown on Petri dishes under 16/8 light/dark at 22 °C (standard condition, SD) for 10 days. Subsequently, one group of WT and mutant plants were maintained under the same conditions for another 10 days, while another group was transferred to continuous white light (WL) at room temperature to grow for another 10 days. The fifth and sixth leaves were harvested and used for detection of cell death and histochemical detection of O2 – at the end of these treatments. Cell death was examined by Evan’s Blue staining as described in Dong et al. (2007). O2 – accumulation in leaves was detected according to Fitzgerald et al. (2004).
Transmission electron microscopy of Arabidopsis chloroplasts
The first true leaf (yellow in cbd) from 10-day-old seedlings (at least 10 seedlings for each) was grown on 1/2 MS + 1% sucrose agar, fixed with 2.5% glutaraldehyde and 2% osmium tetroxide in 0.1M phosphate buffer (pH 7.2) and embedded with resin Eponate 12 after serial dehydration with ethanol to observe chloroplast structure. Ultrathin sections were floated onto 200-mesh copper grids (Ted Pella, Redding, CA, USA) which had been coated with Formvar. Grids were stained for approximately 30 minutes with a 2% solution of uranyl acetate, and then rinsed with distilled water before staining for 10 minutes in Reynolds lead citrate (Reynolds, 1963). Grids were left to dry at least 24 hours before observation in a Philips CM 10 transmission electron microscope (TEM).
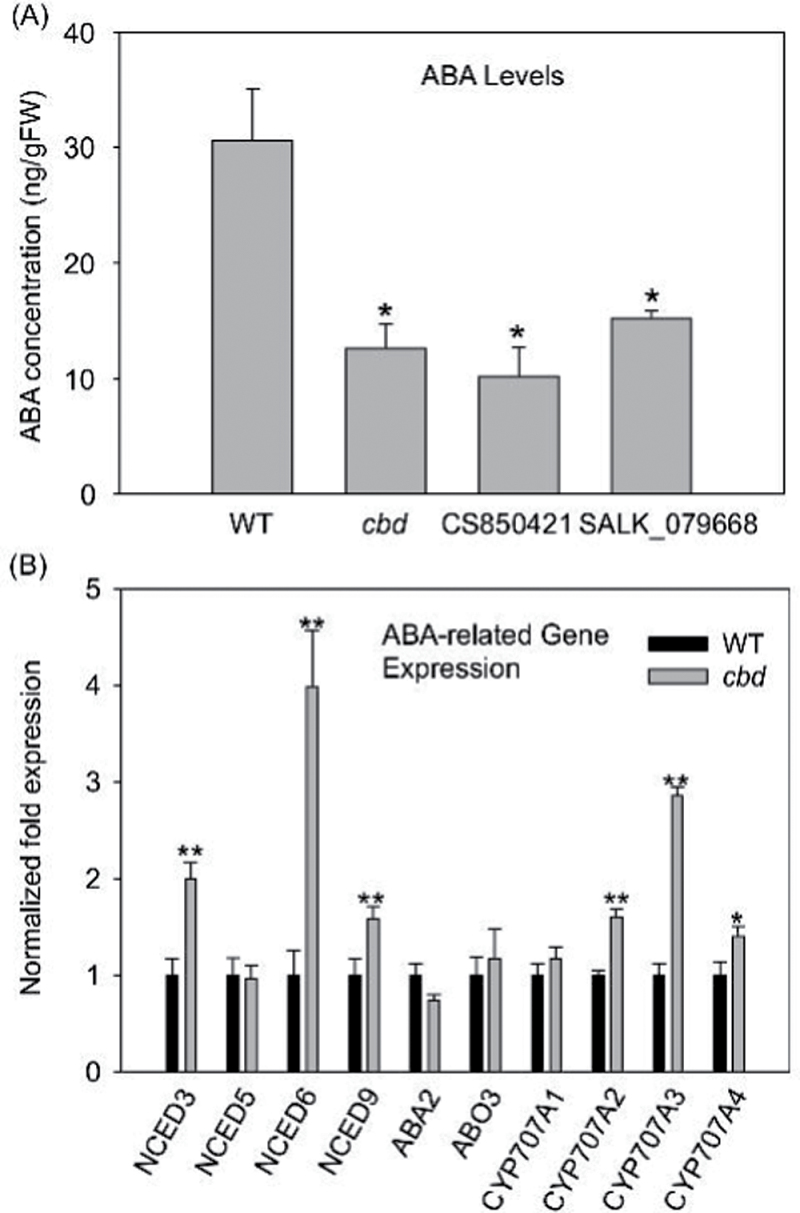
ABA quantification
To quantify the ABA content of cbd, CA850421, SALK_079668, and WT plants, approximately 20mg of respective 7-day-old seedlings was extracted with 80% methanol as described by Galpaz et al. (2008). ABA was quantified by enzyme immunoassay using a Phytodetek ABA test kit (Agdia, Elkhart, IN, USA) following the manufacturer’s instructions. Triplicate samples were used and analysed statistically using ANOVA.
Results
Identification of the cbd mutant
The cbd mutant was identified from the SK A. thaliana T-DNA activation tagged mutant population (Robinson et al., 2009). Seven-day-old seedlings of the cbd mutant had yellow cotyledons (Fig. 1A), which became greener with time. All newly emerged true leaves were yellow and became increasingly green as they matured, but not to the same extent as WT leaves(Fig. 1A, 1B). Stems, inflorescences, flower bud sepals, and siliques were also pale yellow, and all of these organs became light green later in development. Mature cbd mutant plants grew slower and were smaller than WT (Fig. 1C).
Fig. 1.
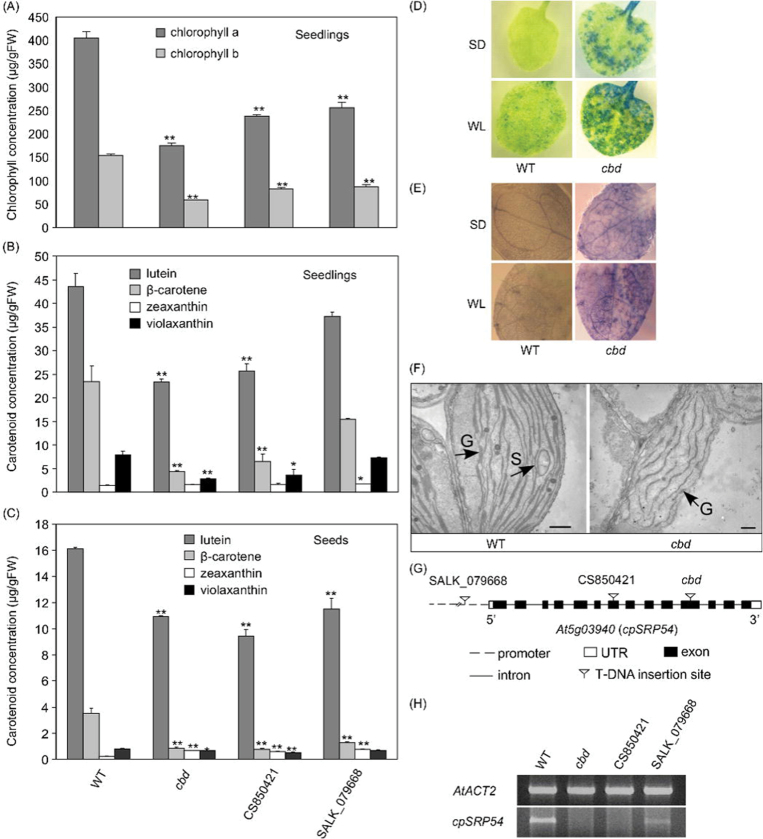
Phenotypes of cbd, SC850421 and SALK_079668 mutants. (A) 7-day-old mutant WT seedlings grown on 1/2 MS media. (B) 25-day-old mutant and WT plants grown in a co-co soil-less mix in the greenhouse. (C) Mature plants of mutants and WT, respectively.
Total chlorophyll was reduced by more than 50% in the 7-day-old seedling of cbd, with both chlorophyll a and b significantly decreased (Fig. 2A). Seedlings of the mutant had approximately 40% of total carotenoid compared to WT. All individual carotenoid compounds were reduced in cbd seedlings except for the minor carotenoid, zeaxanthin, which was not affected (Fig. 2B).The total and individual carotenoid contents of cbd mature seeds were also lower than that of WT seeds, with the exception of zeaxanthin, which was somewhat higher in the mutant (Fig. 2C). To examine if the phenotype in the cbd mutant was caused by a single gene mutation, the mutant line was backcrossed to WT. All F1 progeny (n = 27) had a WT appearance. The segregation ratio of F2 progeny phenotypes (302:107, WT/mutant) indicated that the cbd phenotype is due to a single recessive mutation.
Fig. 2.
Characterization of chloroplasts and cpSRP54 transcript abundance in cpSRP54-deficient mutant lines and WT Arabidopsis. (A) Chlorophyll content of 7-day-old seedlings of mutant lines and WT. (B) Carotenoid content of 7-day-old seedlings of mutant lines and WT. (C) Carotenoid content of mature seeds of mutant lines and WT. Values are mean ± standard error of three extractions from independent seedling plates or seed batches. Significant difference was set at P ≤ 0.05 (*) and P ≤ 0.01 (**) relative to WT Arabidopsis. (D, E) Representative staining of leaves of WT and cbd plants grown under standard light (SD, 16/8 light/dark) and continuous white light (WL): (D) Evan’s Blue, where dead cells show as blue-green patches; (E) nitroblue tetrazolium, where the presence of superoxide results in a purple-blue precipitate. (F) Representative TEM of chloroplasts of WT and cbd. G, grana; S, starch granule. Bar, 500nm. (G) Schematic diagram of T-DNA insertion sites in cpSRP54 mutant lines cbd, CS850421, and SALK_079668. (H) Semi-quantitative RT-PCR analysis of cpSRP54 in mutant lines and WT.
Cell death and superoxide accumulation in cbd leaves
Evan’s Blue stain was used to investigate the extent of cell death in young yellow leaves of the cbd mutant. Under both SD and WL conditions, large blue-green patches of dead cells were observed in cbd leaves (Fig. 2D). More severe cell death occurred under WL in cbd, while little obvious staining was found in WT. The accumulation of superoxide (O2 –), a major species of ROS, was detected by staining the young leaves with nitroblue tetrazolium and observing the purple-blue precipitate. Under SD, superoxide was not detected in WT but it could be easily seen in cbd (Fig. 2E). Furthermore, cbd accumulated abundant superoxide under WL.
Defective chloroplast development in the cbd mutant
Reduced chlorophyll and carotenoid levels and yellow phenotypes in true leaves of the cbd mutant suggested possible defects in chloroplast development. To analyse the effect of the cpSRP54 mutation on chloroplast development, the chloroplast ultra-structure of the first true leaf from 10 individual 10-day-old seedlings of WT and cbd was examined by TEM. Chloroplasts were well developed and organized in the WT mesophyll cells. However, the chloroplasts of cbd had reduced grana stacking with fewer linkages between grana (Fig 2F).
Molecular and genetic characterization of the cbd mutant
TAIL-PCR revealed that a single T-DNA was inserted into the 11th exon of the At5g03940 locus (Fig. 2G). This encodes the 54kDa subunit of the chloroplast signal recognition particle (cpSRP54), also referred to as Fifty Four Chloroplast homologue (FFC). Two mutants, two allelic salk line mutants, SALK_079668 (Alonso et al., 2003) and CS850421, showed morphological phenotypes similar to those of cbd, although not as extreme (Fig. 1). In addition, SALK_079668 and CS850421 had similar chlorophyll and carotenoid patterns in both 7-day-old seedlings and carotenoid patterns in mature seeds (Fig. 2B, 2C), although SALK_079668 was not as affected as CS850421 and cbd.
Gene expression profile in mutants
Semi-quantitative RT-PCR was conducted to analyse cpSRP54 gene expression in cbd mutant and two allelic salk line mutants. SALK_079668 had one T-DNA insertion in the promoter region (–326bp) and CS850421 had one T-DNA in the 7th exon of the At5g03940 locus (Fig. 2G). To show that these two recessive mutants were allelic to cbd, reciprocal crosses between these mutants and cbd were conducted. All the F1 progeny (n = 156 for cbd {female} × CS850421 {male}; n = 123 for CS850421{female} × cbd {male}; n = 47 for cbd {female} × SALK_079668 {male}; n = 14 for SALK_079668 {female} × cbd {male}) showed the same phenotype as cbd. These genetic data confirmed the recessive nature of cbd and that CS850421 and SALK_079668 were allelic to it.
Semi-quantitative RT-PCR was conducted to examine expression of cpSRP54 gene in 7-day-old seedlings of the three mutant plants. Analysis showed that the transcript of the full-length cpSRP54 coding region (CDS) was undetectable in cbd and CS850421 mutants, while SALK_079668 had a much lower level of full-length cpSRP54 transcript compared to WT (Fig. 2H). This is consistent with the observed total carotenoid level for SALK_079668, which although reduced in comparison to WT, was higher than in cbd and CS850421 (Fig. 2B, 2C).
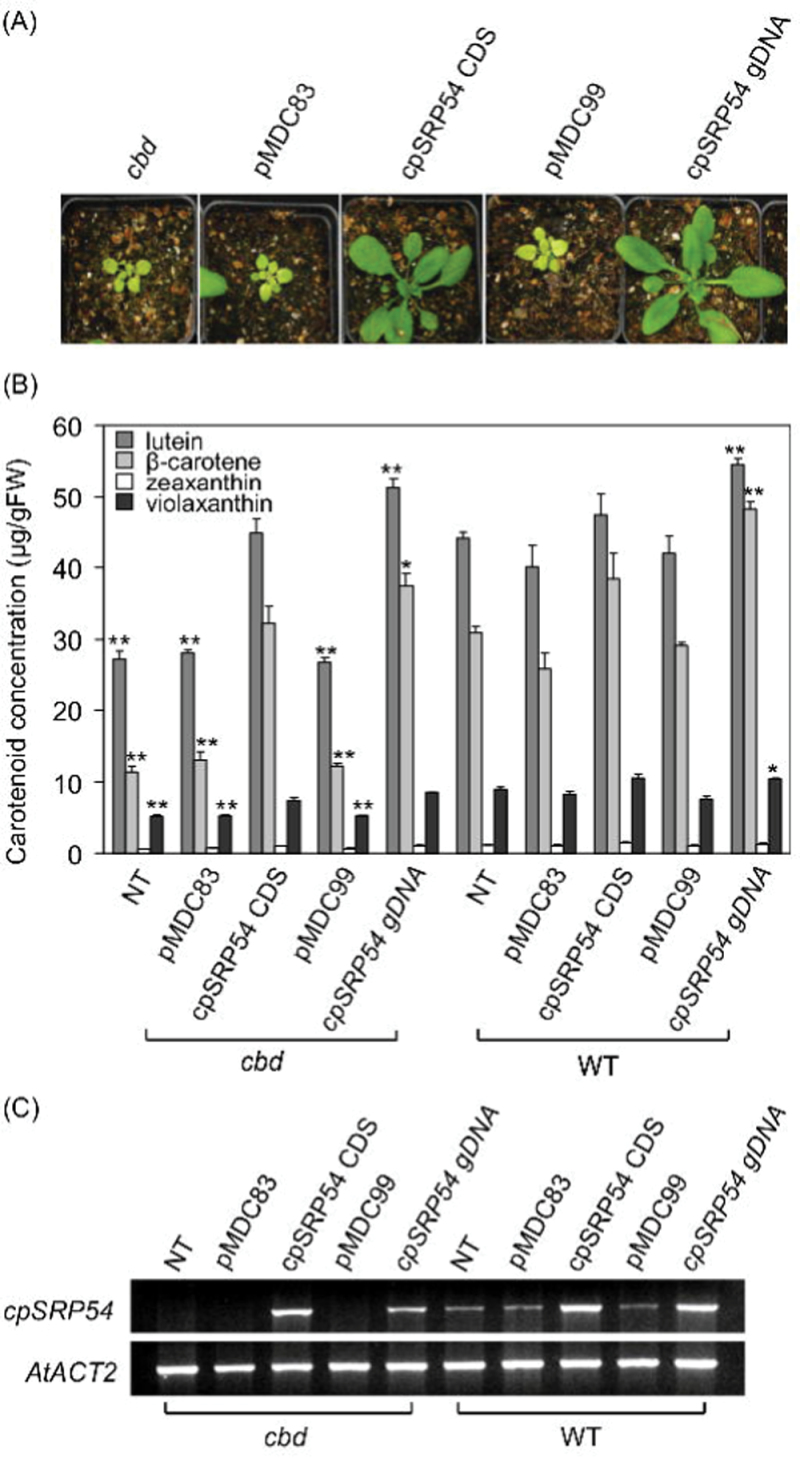
A molecular complementation experiment was conducted to confirm whether the cbd phenotype was caused by a mutation in cpSRP54. Constructs containing cpSRP54 gDNA and CDS were introduced individually into cbd. T2 transgenic plants resulting from both cbd complementation strategies (20 lines per construct) showed restoration of the WT growth and colour phenotype (Fig. 3A). WT levels of carotenoids were shown in seedlings of cbd complementation lines constructed with CDS, but levels were 15% higher in complementation lines constructed with the gDNA relative to WT (Fig. 3B). This carotenoid pattern resulted from cpSRP54 transcript levels (qRT-PCR analysis) which were only slightly higher than WT in cbd plants transformed with the cpSRP54 gDNA, but much stronger in mutant plants transformed with cpSRP54 CDS (potentially due to the strong constitutive promoter, CaMV35S) (Fig. 3C). Both constructs were also used to overexpress cpSRP54 in WT background. Visible phenotype differences were not observed in these over-expression transgenic lines (20 lines per construct) (data not shown). However, the total carotenoid level (lutein and β-carotene) of 8-day-old seedlings transformed with cpSRP54 gDNA was 15% higher than in WT or over-expression lines transformed with CDS, even though transcript levels of the two constructs were relatively similar in this background (Fig. 3B, 3C). Taken together, these data clearly demonstrate that disruption of cpSRP54 was the cause of the altered leaf and cotyledon colour and altered carotenoid profile in the cbd mutant.
Fig. 3.
Complementation of cbd and over-expression of Arabidopsis cpSRP54 in WT Arabidopsis. (A) Representative phenotypes of Arabidopsis cbd lines complemented with an Arabidopsis cpSRP54 gDNA or CDS. Three-week-old seedlings are shown. From left to right: non-transformed cbd, cbd transformed with empty vector pMDC83, cbd with pMDC83-cpSRP54 CDS, cbd with empty vector pMDC99, and cbd with pMDC99-cpSRP54 gDNA. (B) Representative carotenoid content of 8-day-old seedlings. Value are mean ± standard error of three cbd-complemented lines per construct and three over-expression lines per construct. Significant difference was set at P ≤ 0.05 (*) and P ≤ 0.01 (**) relative to WT Arabidopsis. (C) Semi-quantitative RT-PCR confirmation of cpSRP54 expression in cbd-complemented and over-expression transgenic lines as indicated. The CDS of cpSRP54 was amplified and the AtACT2 gene was used as an internal control. NT, non-transformed control.
Impact of cpSRP54 expression changes on the transcription of carotenoid and photosynthesis-related genes
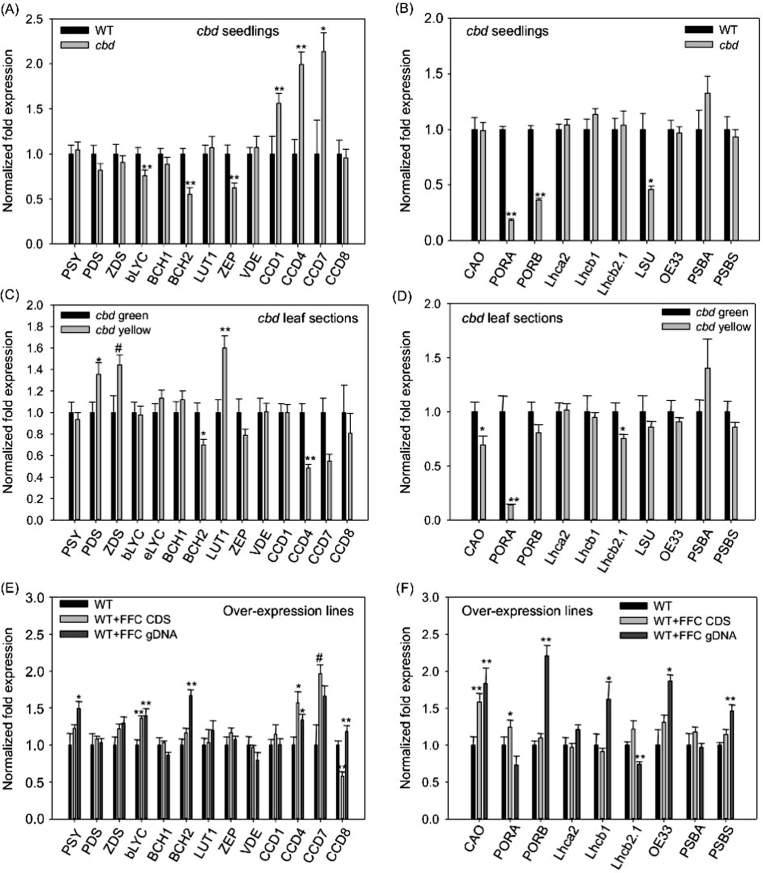
Given the reduced levels of carotenoids and chlorophylls in seedlings of the cbd mutant, qRT-PCR was carried out on 7-day-old seedlings of cbd and WT to examine whether the cpSRP54 mutation had an impact on the expression of carotenoid and chlorophyll biosynthetic genes. Expression of lycopene β-cyclase (bLYC), and especially β-carotene hydroxylase 2 (BCH2) and zeaxanthin epoxidase (ZEP), was significantly decreased in cbd compared to WT (Fig. 4A). Phytoene synthase (PSY), the key enzyme in the carotenoid biosynthesis pathway, and other biosynthetic enzymes did not show transcript differences between cbd and WT; neither did PSY protein levels show changes (Supplementary Fig. S1, available at JXB online). In contrast, carotenoid cleavage dioxygenases (CCD) were oppositely affected such that higher transcript levels were observed for CCD1, CCD4, and CCD7 in cbd seedlings (Fig 4A). Transcripts of both protochlorophyllide oxidoreductase a and b (PORA and PORB) and the chloroplast-encoded large subunit of Rubisco (LSU) were significantly decreased in the leaves of cbd mutant (Fig. 4B). In contrast, transcription of CAO (chlorophyllide a oxygenase), which converts chlorophyllide a to chlorophyllide b, was not significantly affected in the mutant (Fig. 4B), even though chlorophyll b content was decreased. Expression of light-harvesting antenna protein genes for photosystems (PS) I and II (Lhca and Lhcb, respectively), proteins that utilize other thylakoid targeting pathways such as PSBS (spontaneous pathway), OE33 (Sec pathway) and the chloroplast-encoded gene PSBA (D1), which requires cpSRP54 to translocate to its thylakoid location (Amin et al., 1999), were also not affected.
Fig. 4.
Quantitative real-time PCR analysis of carotenoid and photosynthesis gene transcripts in the cbd mutant and cpSRP54 + over-expression Arabidopsis lines. (A) Carotenoid gene profiles and (B) chlorophyll and photosynthesis gene profiles in cbd 7-day-old seedlings. (C) Carotenoid gene profiles and (D) chlorophyll and photosynthesis gene profiles in yellow and green sections of cbd rosette leaves. (E) Carotenoid gene profiles and (F) chlorophyll and photosynthesis gene profiles in 8-day-old seedlings of three Arabidopsis cpSRP54 over-expression lines per construct. Values are mean ± standard error of three replicates. Significant difference was set atP ≤ 0.05 (*) and P ≤ 0.01 (**) relative to WT Arabidopsis (A, B, E, F) and green sections of cbd (C, D).
Gene expression was examined in more detail in green sections relative to yellow sections in cbd mutant leaves (the third and fourth rosette leaf from nine 30-day-old plants). Expression of the BCH2 and CCD4 genes was significantly lower in yellow sections relative to green ones (Fig. 4C). Surprisingly, the expression of PDS and LUT1 genes was significantly higher in the yellow sections and there was a trend towards higher ZDS expression. The chlorophyll biosynthetic genes, CAO and PORA, and light-harvesting gene Lhcb2.1 also had significantly lower expression in yellow sections (Fig. 4D), even though the pattern in whole seedlings (containing a mixture of green and yellow tissue) was less extreme (Fig. 4B).
Finally, expression of genes related to carotenoids and chlorophylls in Arabidopsis plants overexpressing cpSRP54 was analysed. bLYC and CCD4 had higher expression in the cpSRP54 CDS and gDNA over-expression lines relative to WT (Fig. 4E), as did the chlorophyll biosynthetic gene CAO(Fig. 4F). In some cases, enhanced expression occurred only with gDNA over-expression, e.g. transcripts of PSY, BCH2, and CCD8 were higher only in the cpSRP54 gDNA lines (Fig. 4E). Oddly, CCD8 was reduced in the CDS lines. Higher Lhcb1 and PORB and lower Lhcb2.1 transcripts accumulated in the gDNA over-expression lines (Fig. 4E, 4F) compared to WT. Moreover, transcripts encoding chloroplast proteins utilizing the non-SRP thylakoid targeting pathways, e.g. PSBS and OE33, were significantly higher in the over-expression lines transformed with the gDNA relative to WT and relative to lines overexpressing the cpSRP54 CDS (Fig. 4F).
Plastid-to-nucleus retrograde signalling may regulate nuclear gene expression in cbd
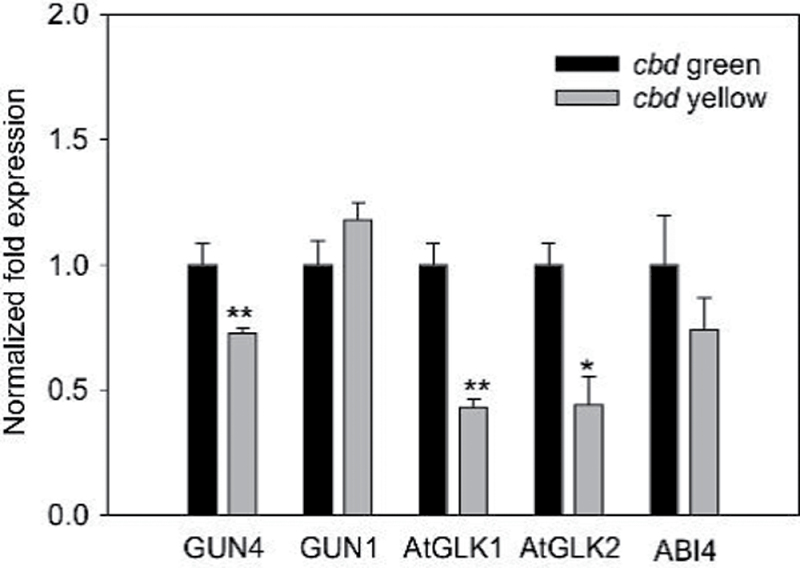
The downregulation of chlorophyll and carotenoid biosynthetic genes in the cbd mutant could be an indirect response to defects in chloroplast development and suggested the potential involvement of a plastid-to-nucleus retrograde signalling mechanism. qRT-PCR showed that the expression of GUN4, but not GUN1, was approximately 30% lower in the yellow sections of cbd than in green sections (Fig. 5). This result suggested that Mg-ProtoIX synthesis could be reduced in the cbd yellow sections. Therefore, it seemed unlikely that downregulation of chlorophyll and carotenoid biosynthetic genes in the cbd yellow sections resulted from the build-up of Mg-ProtoIX. However, AtGLK1 and AtGLK2, transcripts coding for functionally redundant proteins that positively regulate photosynthesis-related nuclear gene expression (PRNGE) in plastid-to-nuclear signalling (Fitter et al., 2002), were decreased 2-fold in yellow sections relative to green sections of the cbd mutant (Fig. 5). Coincidently, the expression of ABI4, which acts as a negative regulator of PRNGE and is part of the GUN1/GUN4 pathway (Koussevitzky et al., 2007), was not changed in yellow leaf sections compared to green sections of the cbd mutant (Fig. 5).
Fig. 5.
Expression patterns of genes involved in the plastid-to-nucleus retrograde signalling in yellow and green sections of the cbd mutant. Significant difference was set at P ≤ 0.05 (*) and P ≤ 0.01 (**) relative to green sections of cbd mutant.
ABA biosynthesis is altered in the cbd mutant
Since carotenoids are precursors of ABA and an increase in CCD transcripts and a reduction of carotenoids in cbd were observed, changes to levels of ABA were anticipated. To test this hypothesis, the ABA content of 7-day-old seedlings of the mutants and WT were measured. The results showed that cbd and the two allelic lines CS850421 and SALK_079668 had 2–3-fold lower ABA levels (Fig. 6A). To investigate whether the cbd mutant has an altered response to ABA, the effect of exogenous ABA on root growth was tested. cbd, the two allelic mutant lines, and WT plants had similar growth kinetics when treated with ABA (data not shown), suggesting that these mutants have a normal response to ABA. Since ABA level was reduced in cbd, the expression of genes involved in ABA metabolism was examined by qRT-PCR. Nine-cis-expoxycarotenoid dioxygenases (NCEDs), the first family of enzymes committed specifically to ABA synthesis, cleave the epoxycarotenoid precursor to form xanthoxin (Nambara and Marion-Poll, 2005). The transcript levels of three of these genes, NCED3, NCED6, and NCED9, were upregulated in cbd (Fig. 6B), whereas those of NCED5, ABA2 (short-chain alcohol dehydrogenase), and AAO3 (abscisic aldehyde oxidase, catalysing the final step in ABA biosynthesis) were not significantly affected in cbd (Fig. 6B). Hence altered ABA biosynthesis was not the cause of the lower ABA level in cbd. The major catabolic route leading to 8'-hydroxy ABA is catalysed by the cytochrome P450 enzyme, ABA 8'-hydroxylase, encoded by members of the CYP707A family, CYP707A1–CYP707A4 (Kushiro et al., 2004). The expression of CYP707A1 and CYP707A2 was significantly increased in the cbd mutant (Fig. 6B), which was consistent with the decreased ABA content.
Fig. 6.
ABA content and expression of ABA metabolic genes in the cbd mutant and WT. (A) ABA content in 7-day-old seedlings. (B) Expression patterns of ABA biosynthetic and catabolic genes in 7-day-old seedlings. Significant difference was set at P ≤ 0.05 (*) and P ≤ 0.01 (**) relative to WT Arabidopsis.
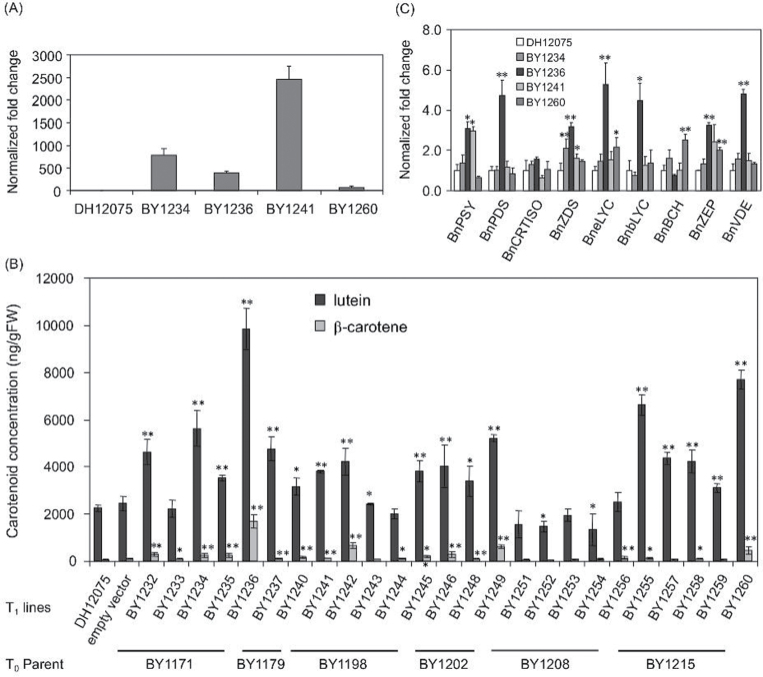
Expression of Arabidopsis cpSRP54 resulted in higher carotenoid accumulation in B. napus seeds
Since over-expression of cpSRP54 elevated carotenoids in Arabidopsis seedlings, a decision was made to determine if cpSRP54 could be used to enhance the carotenoid levels in seeds of the oilseed crop, B. napus. T1 cpSRP54 + developing seeds in a DH12075 background showed varied transcript abundance for cpSRP54 at 34 DPA (Fig. 7A). Growth was not affected in these transgenic B. napus lines; neither was the seed yield changed at all (Supplementary Fig. S2). A range of T0 and T1 transgenic seeds accumulated substantially greater levels of the major B. napus seed carotenoids, lutein and β-carotene, ranging 1.3–10.9-fold higher compared with levels observed in WT DH12075 and empty vector control plants (Fig. 7B for seed from T1 lines; Supplementary Fig. S3 for T0 parental plants). Violaxanthin and zeaxanthin were at trace levels in T1 lines. The T0 plant BY1179 accumulated the highest amount of lutein (9-fold higher than control plants), and 41-fold higher β-carotene occurred in T0 plants BY1179 and BY1198 (Supplementary Fig. S3). Six independent events (plants) with significantly higher total carotenoid, along with one empty vector control line and a WT DH12075 control line, were selected for further analysis. T1 seeds derived from plant BY1171 had the highest amount of lutein (5-fold higher) and β-carotene (10-fold higher) compared with control seed, while levels in T1 plants derived from BY1179 were only 2-fold higher (Fig. 7B). Except for T1 lines derived from BY1208 (BY1251–BY1254), seeds of all other T1 lines had significantly higher (or a trend towards higher) β-carotene and/or lutein. However, transcript levels of cpSRP54 did not correlate well with carotenoid content in the transgenic B. napus seeds.
Fig. 7.
B. napus expressing the Arabidopsis cpSRP54 gene. (A) Semi-quantitative RT-PCR analysis of cpSRP54 transgene expression in T1 developing seeds at 34 days post anthesis (DPA). (B) Carotenoid profiles of B. napus seeds from cpSRP54 + expression lines (T1), an empty vector control plant, and WT DH12075. [Violaxanthin and zeaxanthin were at trace levels.] (C) Carotenoid gene expression profiles in T1 transgenic cpSRP54 + B. napus developing seeds at 34 DPA. Values are mean ± standard error of three replicates. Significant difference was set at P ≤ 0.05 (*) and P ≤ 0.01 (**) relative to DH12075.
Next, a wide range of carotenoid biosynthetic genes in developing seeds of T1 lines were tested at 34 DPA for transcript levels in the cpSRP54 + transgenic B. napus. Gene expression for many of these genes was enhanced relative to control (Fig. 7C) and was consistent with carotenoid content, except for line BY1234. Expression of carotenoid biosynthetic genes was especially enhanced in T1 line BY1236, which is consistent with its highest carotenoid content. However, the transcript level of carotenoid isomerase (CRTISO) was not significantly changed in any lines tested.
To investigate whether altered carotenoid profiles affected fatty acid biosynthesis in the seeds of transgenic lines, oil content and fatty acid (FA) profiles were measured in T0 and T1 seeds (Supplementary Table S1). Of the 22 T0 independent transgenic plants tested, the majority (14 lines) had FA profiles and oil content identical to DH12075 control seed. Only eight had modified FA profiles and lower oil content, ranging from 26.13% freshweight (FW) in BY1179 seeds to 39.33% in line BY1180. Five lines with reduced oil content were detected in the T1 generation as well, and the lines with reduced oil content included those with increased carotenoid levels. Increased unsaturated fatty acids C18:2 and C18:3 and decreased C18:1 were also shown in some lines over both generations compared to DH12075 and empty vector control plants, for example T0 plants BY1170 and BY1202 and T1 lines BY1241 and BY1260. Overall, however, the magnitude of these changes to fatty acids was small or non-existent.
Discussion
In addition to their many benefits to human health and nutrition, carotenoids are essential components of the plant light-harvesting system and play significant roles in plant biology. These roles include photosynthesis, protection against photooxidative damage, and membrane stabilization (Botella-Pavía and Rodríguez-Concepción, 2006). Therefore, a major deficiency in carotenoid biosynthesis or accumulation, such as occurred with the current Arabidopsis cbd mutant, would be expected to affect plant development and chloroplast assembly. Dong et al. (2007) reported that A. thaliana carotenoid deficient mutants spc1-1 (spontaneous cell death1-1) and spc1-2, which have mutations in the gene encoding a putative ζ-carotene desaturase (ZDS), had bleached leaves, excessive production of superoxide radicals, abnormal chloroplast development, and arrest of growth leading to seedling lethality. The current cbd mutant, with mutation in cpSRP54, manifested several similar phenotypes similar to those of spc mutants: reduced carotenoid and chlorophyll content in the seedlings, yellow cotyledons and first true leaves, as well as reduced growth. Molecular complementation of cbd, using both genomic and cDNA sequences, confirmed that the phenotypic changes were due to the mutational disruption of cpSRP54.
Over-expression of a genomic copy of cpSRP54 in WT Arabidopsis led to an increase in total carotenoid content without visible morphological or developmental effects. However, lines transformed with the cpSRP54 CDS, in spite of much higher levels of cpSRP54 transcript than WT, did not show a significantly altered total carotenoid content. This suggests that more complex regulatory mechanisms may impact cpSRP54 expression, such as post-transcriptional gene silencing (Stam et al., 1997) or RNA splicing, which could result in altered and non-functional enzymes or post-translational modification. A similar finding was reported for the N plant resistance gene where transgenic plants expressing the N cDNA failed to exhibit complete resistance to tobacco mosaic virus, whereas transgenic plants harbouring a cDNA bearing an intron and containing 3'-N genomic sequences were resistant (Dinesh-Kumar and Baker, 2000). This indicates that for some genes, expression of genomic sequences may be preferable to cDNA to achieve complete protein functionality.
Carotenoids are essential for quenching excessive free radicals and reactive oxygen species (ROS) and providing protection against photooxidation (Botella-Pavía and Rodríguez-Concepción, 2006). Plants with reduced carotenoids are known to generate photooxidative species such as ROS (Dong et al., 2007). Due to the substantial reduction of carotenoids in the Arabidopsis cbd mutant, an enhancement of superoxide could be detected in this mutant. This and potentially other ROS molecules could be the reason for the slow growth, small stature, and leaf cell death phenotype in this mutant. The carotenoid deficiency mutant spc1, encoding a ZDS essential for carotenoid biosynthesis, also showed defects in chloroplast development (Dong et al., 2007). The cbd chloroplast had reduced grana thylakoids, indicating the importance of cpSRP54 for plastid development and plant growth. This is consistent with a study on over-expression of dominant negative forms of cpSRP54, in which the alteration of a single amino acid in Arabidopsis resulted in delayed chloroplast development in the first true leaf with fewer thylakoid membranes and grana stacks than in control counterparts (Pilgrim et al., 1998).
The cbd mutant showed reduced amounts of total carotenoids with very few changes in the transcription of genes encoding carotenoid biosynthetic enzymes. Although BCH2 expression was decreased in cbd, its related minor carotenoid, zeaxanthin, showed no substantial change in level. This inconsistency could be due to redundant function of BCH1 and/or lower expression of ZEP. The cbd mutant and allelic SALK lines also had similar levels of PSY protein relative to WT. This suggests that mutation of cpSRP54 did not substantially affect the expression or translation of carotenoid biosynthetic genes. Nevertheless, the expression of several CCD, NCEDs and ABA-related CYP450 genes were enhanced in cbd. The CCD1 enzyme cleaves a variety of carotenoid substrates at the C9–10 and C9'-10' double bonds (Schwartz et al., 2001). CCD4 can participate in dark-induced breakdown of carotenoids (Ytterberg et al., 2006) and CCD7 coordinately synthesizes strigolactones with CCD8 (Vogel et al., 2010). This upregulation of the CCD and NCED genes could lead to increased catabolism of carotenoids and hence to reduced carotenoids in cbd. Strangely, expression of NCEDs, encoding enzymes committed specifically to ABA synthesis, was upregulated in cbd (Fig. 6B). However, the increase in expression of ABA catabolic genes, CYP707A family members, is consistent with the decreased ABA content found in the cbd mutant.
A plastid-to-nucleus retrograde signalling mechanism has been reported to regulate the expression of photosynthetic genes in the nucleus, and Lhcb is perhaps most responsive to this signal (Strand et al., 2003). In the current study, there was evidence suggesting disruption of this chloroplast retrograde signalling in the cbd mutant by a decrease in transcripts of AtGLK1, AtGLK2, and the retrograde signalling gene, GUN4 in yellow sections of the cbd mutant. Reduced gene expression of Lhcb2.1 and PORA in yellow leaf sections and the lower expression of the chlorophyll biosynthetic genes PORA, PORB, and LSU in whole seedlings could be a response to this change in plastid-to-nucleus signalling pathway. The increase in expression of a few carotenoid biosynthetic genes and the decrease in expression of carotenoid degradation genes in yellow leaf sections of the cbd mutant suggest that the cbd mutant is attempting to increase the antioxidation capacity in the leaf. Thus AtGLK1 and AtGLK2 may be involved in fine-tuning carotenogenesis and degradation in response to the functional state of the plastid. However, this does not preclude the possibility that plastid retrograde signalling may be disrupted by enhanced levels of ROS resulting from reduced carotenoid accumulation (Foudree et al., 2010).
The storage and sequestration of carotenoids within various plastid types is an important regulatory mechanism for carotenoid accumulation. Organelle biogenesis partly determines the size of the carotenoid storage compartment of plastids (Lopez et al., 2008; Cazzonelli and Pogson, 2010) and carotenoids co-accumulate with chlorophyll in a pigment-binding protein complex embedded in thylakoid membranes in leaf tissues (Botella-Pavía and Rodríguez-Concepción, 2006). Both hp-2 and hp-3 mutants had enlarged plastid compartment sizes, which were coupled with enhanced levels of carotenoid (Kolotilin et al., 2007; Galpaz et al., 2008). It also has been reported that the carotenogenic pathway is appreciably stimulated in the presence of sequestering structures (Rabbani et al., 1998). Consequently, the defects of chloroplast development in cbd and, hence, the reduced storage compartment size for carotenoid accumulation in this mutant, could contribute to the decreased carotenoid content in cbd. Impaired chloroplast targeting could also feedback via the plastid-to-nucleus retrograde signalling to reduce chlorophyll and carotenoid content in cbd seedlings.
All true leaves of the cbd mutant were yellow during early stages of development but became greener as they matured, Moreover, cbd could survive in the homozygous state, suggesting that the mutation is not lethal and that alternate thylakoid protein targeting systems can compensate for the loss of the cpSRP pathway, depending on the developmental stage. This possibility is supported by the work of Tzvetkova-Chevolleau et al. (2007), who showed that cpSRP43 functions independently of cpSRP54/cpFtsY when targeting LHCPs to the thylakoid membrane. These observations are consistent with the idea that cpSRP54 is non-essential and that plants can compensate for its loss by employing alternate thylakoid targeting pathways.
Suppression of cpSRP54 expression appears to unleash a cascade of molecular and physiological events with consequences on plant growth and development in the Arabidopsis cbd mutant. Disruption of the plastid-to-nucleus retrograde signalling and deformed plastids may be responsible in part for the reduced carotenoid content in this line. Reduced carotenoid content impacts the antioxidative capacity of the cell, thus allowing for the accumulation of ROS, which affects plant performance as manifested by reduced growth, stunted appearance, and enhanced cell death. On the other hand, over-expression of cpSRP54 in Arabidopsis and expression in B. napus enhances seed carotenoid content, with no significant impact on growth, seed oil, or seed yield. These experiments point to the Arabidopsis cpSRP54 gene as a new tool/mechanism for enhancing carotenoids and improving the nutritional value of crop plants. Increased carotenoid levels have also been enhanced in another important crop (potato tubers) by inducing the formation of chromoplasts containing carotenoid-sequestering structures using the Or transgene (Lopez et al., 2008).
Supplementary Material
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Western blot analysis of PSY extracted from 7-day-old seedlings of cbd mutant and wild-type Arabidopsis
Supplementary Fig. S2. Seed yield of T0 and T1 B. napus plants expressing Arabidopsis cpSRP54
Supplementary Fig. S3. Carotenoid profiles of B. napus seeds from cpSRP54 - expression lines (T0), an empty vector control plant, and wild-type DH12075
Supplementary Table S1. Oil content and fatty acid profiles of T0 and T1 B. napus seed expressing Arabidopsis cpSRP54
Acknowledgements
The authors thank Dr. Guosheng Liu for technical assistance with TEM and the ABRC for distributing Arabidopasis mutant seeds SALK_079668 and CS850421. Funding was provided by Genome Alberta/Genome Canada as part of the ‘Designing Oilseeds for Tomorrow’s Markets’ project.
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Amin P, Sy DAC, Pilgrim ML, Parry DH, Nussaume L, Hoffman NE. Arabidopsismutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant physiology. 1999;121:61–70. doi: 10.1104/pp.121.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty S, Nolan J, Kavanagh H, O’Donovan O. Macular pigment optical density and its relationship with serum and dietary levels of lutein and zeaxanthin. Archives of Biochemistry and Biophysics. 2004;430:70–76. doi: 10.1016/j.abb.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Botella-Pavía P, Rodríguez-Concepción M. Carotenoid biotechnology in plants for nutritionally improved foods. Physiologia Plantarum. 2006;126:369–381. [Google Scholar]
- Cazzonelli CI, Pogson BJ. Source to sink: regulation of carotenoid biosynthesis in plants. Trends in Plant Science. 2010;15:266–274. doi: 10.1016/j.tplants.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D. Carotenoid synthesis and function in plants: insights from mutant studies in Arabidopsis. Pure and Applied Chemistry. 1999;71:2205–2212. [Google Scholar]
- Dinesh-Kumar SP, Baker BJ. Alternatively spliced Nresistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proceedings of the National Academy of Sciences, USA. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Deng Y, Mu J, Lu Q, Wang Y, Xu Y, Chu C, Chong K, Lu C, Zuo J. The Arabidopsis Spontaneous Cell Death1gene, encoding a ζ-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Research. 2007;17:458–470. doi: 10.1038/cr.2007.37. [DOI] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. The Plant Journal. 2002;31:713–727. doi: 10.1046/j.1365-313x.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald HA, Chern M-S, Navarre R, Ronald PC. Overexpression of (At)NPR1in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Molecular Plant–Microbe Interactions. 2004;17:140–151. doi: 10.1094/MPMI.2004.17.2.140. [DOI] [PubMed] [Google Scholar]
- Foudree A, Aluru M, Rodermel S. PDS activity acts as a rheostat of retrograde signaling during early chloroplast biogenesis. Plant Signaling & Behavior. 2010;5:1619–1622. doi: 10.4161/psb.5.12.13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Takita E, Harada H, Sakurai N, Suzuki H, Ohyama K, Shibata D, Misawa N. Pathway engineering of Brassica napusseeds using multiple key enzyme genes involved in ketocarotenoid formation. Journal of Experimental Botany. 2009;60:1319–1332. doi: 10.1093/jxb/erp006. [DOI] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J. Abscisic acid deficiency in the tomato mutant high-pigment 3leading to increased plastid number and higher fruit lycopene content. The Plant Journal. 2008;53:717–730. doi: 10.1111/j.1365-313X.2007.03362.x. [DOI] [PubMed] [Google Scholar]
- Hannoufa A, Hossain Z. Regulation of carotenoid accumulation in plants. Biocatalysis and Agricultural Biotechnology. 2012;1:198–202. [Google Scholar]
- Howitt C, Pogson BJ. Carotenoid accumulation and function in seeds and non-green tissues. Plant, Cell and Environment. 2006;29:435–445. doi: 10.1111/j.1365-3040.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- Inskeep WP, Bloom PR. Extinction coefficients of chlorophyll a and b in N N-dimethylformamide and 80% acetone. Plant Physiology. 1985;77:483–485. doi: 10.1104/pp.77.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Che F-S, Terauchi R, Inaba T. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signalling. Plant Physiology. 2009;151:1339–1353. doi: 10.1104/pp.109.145987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolotilin I, Koltai H, Tadmor Y, Bar-Or C, Reuveni M, Meir A, Nahon S, Shlomo H, Chen L, Levin I. Transcriptional profiling of high pigment-2dgtomato mutant links early fruit plastid biogenesis with its overproduction of phytonutrients. Plant Physiology. 2007;145:389–401. doi: 10.1104/pp.107.102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. The Arabidopsiscytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism. The EMBO Journal. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. GUN4, a regulator of chlorophyll synthesis and intracellular signalling. Science. 2003;299:902–906. doi: 10.1126/science.1079978. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thalianaT-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal. 1995;8: 457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Lopez AB, Eck JV, Conlin BJ, Paolillo DJ, O’Neill J, Li L. Effect of the cauliflower Ortransgene on carotenoid accumulation and chromoplast formation in transgenic potato tubers. Journal of Experimental Botany. 2008;59:213–223. doi: 10.1093/jxb/erm299. [DOI] [PubMed] [Google Scholar]
- McCarty DR. Genetic control and integration of maturation and germination pathways in seed development. Annual Review of Plant Physiology and Plant Molecular Biology. 1995;46:71–93. [Google Scholar]
- Michiels A, Van den Ende W, Tucker M, Van Riet L, Van Laere A. Extraction of high-quality genomic DNA from latex-containing plants. Analatytical Biochemistry. 2003;315:85–89. doi: 10.1016/s0003-2697(02)00665-6. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Nott A, Jung H-S, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signalling. Annual Review of Plant Biology. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Pilgrim ML, van Wijk K-J, Parry DH, Sy DAC, Hoffman NE. Expression of a dominant negative form of cpSRP54 inhibits chloroplast biogenesis in Arabidopsis. The Plant Journal. 1998;13:177–786. doi: 10.1046/j.1365-313x.1998.00021.x. [DOI] [PubMed] [Google Scholar]
- Pogson BJ, McDonald K, Truong M, Britton G, DellaPenna D. Arabidopsiscarotenoid mutants demonstrate lutein is not essential for photosynthesis in higher plants. The Plant Cell. 1996;8:1627–1639. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani S, Beyer P, Lintig JV, Hugueney P, Kleinig H. Induced β-carotene synthesis driven by triacylglycerol deposition in the unicellular alga Dunaliella bardawil. Plant Physiology. 1998;116:1239–1248. doi: 10.1104/pp.116.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask L, Ellerstrom M, Ezcurra I, Stalberg K, Wycliffe P. Seed-specific regulation of the napin promoter in Brassica napus. Journal of Plant Physiology. 1998;152:595–599. [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. Journal of Cell Biology. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SJ, Tang LH, Mooney BA, et al. An archived activation tagged population of Arabidopsis thalianato facilitate forward genetics approaches. BMC Plant Biology. 2009;9:101. doi: 10.1186/1471-2229-9-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JAD. Characterization of a novel carotenoid cleavage dioxygenase from plants. Journal of Biological Chemistry. 2001;267:25208–25211. doi: 10.1074/jbc.M102146200. [DOI] [PubMed] [Google Scholar]
- Stam M, Mol J, Kooter J. The silence of genes in transgenic plants. Annals of Botany. 1997;79:3–12. [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication trigged by accumulation of Mg-protoporphyrinIX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kawazu T, Koyama H. RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. BioTechniques. 2004;37:542–544. doi: 10.2144/04374BM03. [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T, Hutin C, Noël LD, et al. Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. The Plant Cell. 2007;19:1635–1648. doi: 10.1105/tpc.106.048959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeno D, Tobias AV, Arnold FH. Diversifying carotenoid biosynthetic pathway by directed evolution. Microbiology and Molecular Biology Reviews. 2005;69:51–78. doi: 10.1128/MMBR.69.1.51-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Walter MH, Giavalisco P, et al. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. The Plant Journal. 2010;61:300–311. doi: 10.1111/j.1365-313X.2009.04056.x. [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier J-B, van WiJk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiology. 2006;140:984–999. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Lydiate DJ, Schäfer UA, Hannoufa A. Characterization of a β-carotene hydroxylase of Adonis aestivalisand its expression in Arabidopsis thaliana. Planta. 2007;226:181–192. doi: 10.1007/s00425-006-0455-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.