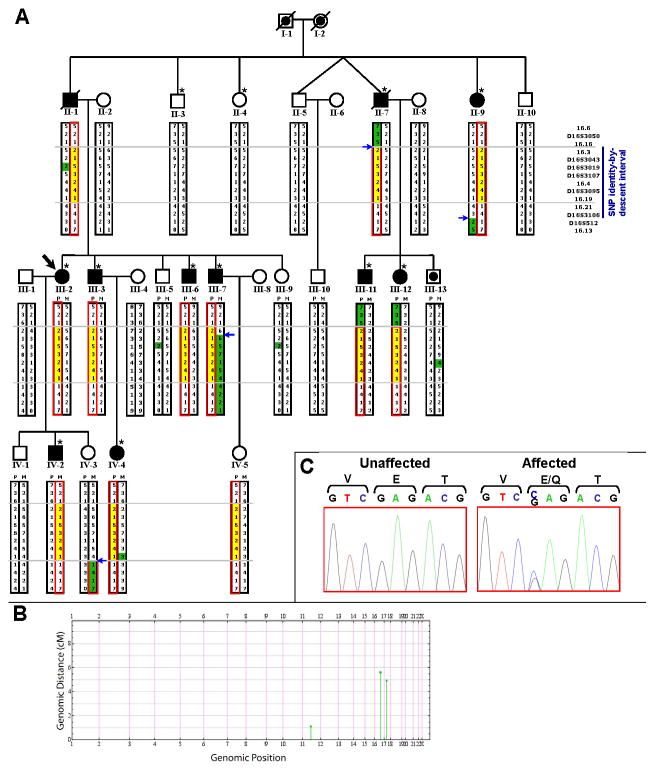
Figure 2. Pedigree with chromosome 16q21-22.1 haplotypes, and NOL3 mutation.
A) Each microsatellite allele for a given marker is denoted by a different integer value. The red box encloses the mutant haplotype. The yellow box encloses the critical region of the mutant haplotype. Blue arrows denote recombination events. Alleles are highlighted in green if they differ from the remainder of that haplotype, because of either a recombination or non-Mendelian inheritance (II-1 and progeny, III-13, IV-4; likely due to spontaneous repeat contraction/expansion). II-1 and II-7 are now deceased but were examined prior to death. IV-3 was an asymptomatic adult with normal SSEP. The dot (III-13) denotes unknown phenotype; III-13 refused SSEP. IV-5 was an asymptomatic 26-year-old at exam (refused SSEP), and is predicted to be a carrier although incomplete penetrance is possible. Forty-two relatives were omitted to preserve anonymity. Asterisks denote subjects who were genotyped for genome-wide SNP identity-by-descent mapping.
B) Genome-wide SNP mapping identified 3 candidate linkage regions (green). IBD block size (cM) is plotted against genomic position. Blocks that were not identical-by-descent have been omitted.
C) Sanger-sequencing chromatographs confirmed co-segregation of the NM_001185058.1:c.61G>C variant, predicted to cause an E21Q mutation in NOL3.