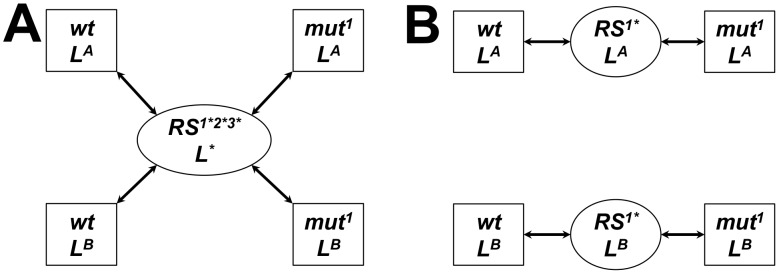
Figure 1. Alchemical thermodynamic paths using SRMM (A) and SRSM (B) in the bound state between wild type (wt) and a mutant (mut1).
The paths (arrows) between end states (squares) going through nonphysical reference states (ovals) are shown. The SRMM uses a common reference state ‘hub’ for all mutations and ligands (RS1*2*3* L*); SRSM uses a mutation and ligand-specific reference state (RS1* LX). Decoupled residues and ligands are noted by an ‘*’. Thermodynamic paths in the unbound state have a similar form but without the ligand.