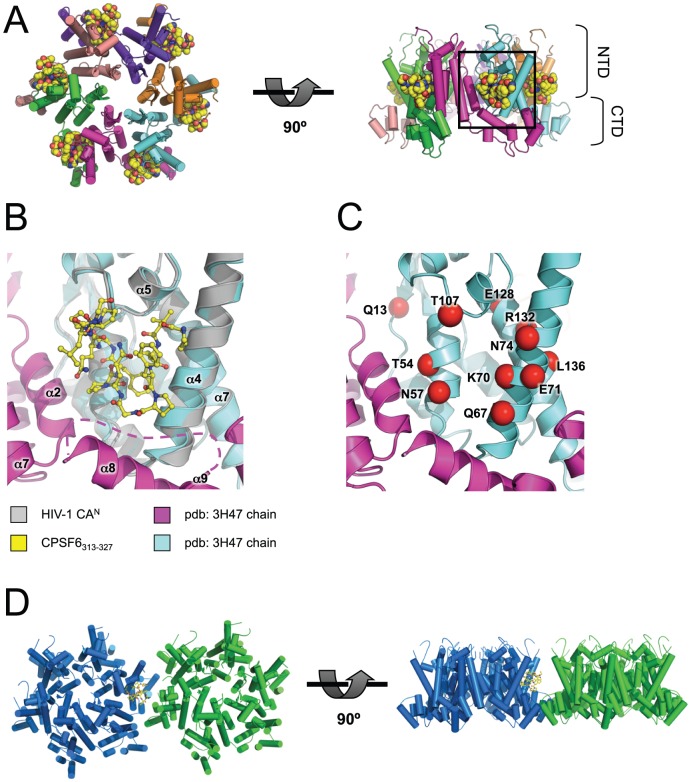
Figure 5. The CPSF6-binding interface is accessible and highly conserved in HIV-1 virions.
(A) Model of CPSF6313–327 binding to HIV-1 CA hexamer. The hexamer structure was derived from pdb: 3H47 [15] by generating symmetry-related copies in PyMOL. The model was composed by superposition of CAN chains from HIV-1 CAN:CPSF6313–327 on the hexamer using secondary structure matching. HIV-1 CA helices are shown as cylinders and CPSF6313–327 as spheres. Left: top view, right: side view. N-terminal and C-terminal CA domains (NTD and CTD) are labeled. (B) Close-up of boxed region in (A). CAs are shown as cartoons and CPSF6313–327 as sticks. The region between CTD positions 175 and 188 was disordered and so is represented by a dashed line. (C) Same view as in (B), showing exposed CA mutations (labelled and shown as red spheres) that result in decreased HIV-1 infectivity [14] (see Figure 1 ). (D) Model of CPSF6313–327 binding at a hexamer-hexamer interface. Neighbouring hexamers were derived from pdb: 3H47 by generating extended symmetry-related copies in PyMOL. HIV-1 CAN:CPSF6313–327 was superposed on the hexamer using secondary structure matching. The model shows that CPSF6313–327 binding is likely to be accommodated at neighbouring CTD-mediated hexamer-hexamer junctions.