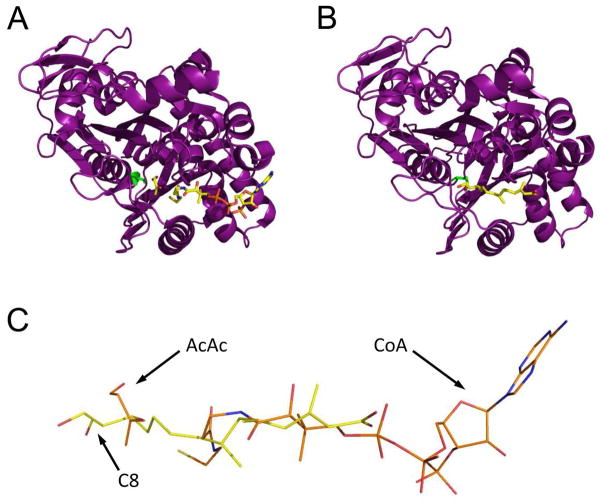
Figure 5.
Comparison of E. faecalis mvaS structures bound to either acetoacetate and CoA or hymeglusin. (A) Chain B from the structure of mvaS bound to acetoacetate and CoA (PDB code 1YSL). The polypeptide is shown in purple cartoon while the ligands are shown in ball and stick format. The catalytic cysteine residue is drawn in green. (B) Chain A from the structure of mvaS bound to hymeglusin (PDB code 3V4X). (C) Superposition of the CoA and hymeglusin structures as they lie within the mvaS active site. Note that the alphatic tail of hymeglusin largely occupies the same position as the pantothenic acid moiety of CoA. The positions of acetoacetate (AcAc), Coenzyme-A (CoA), and the C8 carbon of hymeglusin (C8) are indicated with arrows.