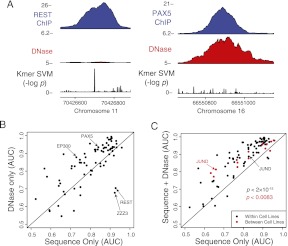
Figure 4.
Combining chromatin and sequence models improves binding site prediction. (A) Binding sites for REST and PAX5 illustrate loci that have a high sequence signal or DNase accessibility, but not both. (B) Learning sequence models in a single cell type reveals that some TFs are better predicted by sequence signals (such as REST), whereas others are better predicted by DNA accessibility (such as EP300 and PAX5). The AUC was determined for each replicate in each cell type and then averaged. (C) When DNase accessibility information is added to k-mer SVM models, the combined model is more predictive of in vivo binding sites. The scatter plot compares the accuracy of a combination of sequence and DNase SVM signatures with that of the sequence model alone. Models were learned from one cell type and then used to predict binding sites in the same cell type (black) or a different cell type (red). Accuracy (AUC) for each TF was averaged across replicates and cell lines (same cell case) or only replicate experiments (transfer learning case). JUND is an outlier, where applying the sequence model across cell lines is significantly worse than applying it in the same cell line. POLR3 is poorly predicted in all settings and is not shown.