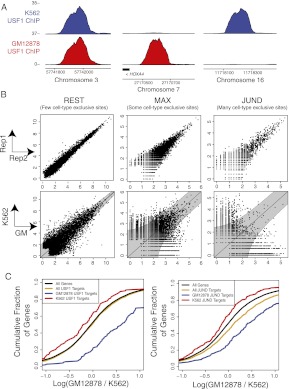
Figure 5.
Cell-type–specific transcription factor binding is measured by ChIP-seq and correlated with differential gene expression. (A) ChIP-seq of USF1 reveals sites that are bound in both cell lines (left), only GM12878 (middle), or only K562 (right). Units are reads per million aligned (RPM). (B) We find cell-type–specific binding sites by measuring replicate-to-replicate noise and comparing it to cell-to-cell variation. Replicate and cell-specific binding are shown for REST, MAX, and JUND. The top row of scatterplots shows the ChIP-seq read counts [in RPM, scaled by log(x + 1)] for the top 5000 peaks in two replicate experiments in the same cell type (GM12878). The bottom row of scatterplots shows the log ChIP-seq read counts in GM12878 versus K562 for the union of the top 5000 peaks in each cell line. In these plots, each point is a binding site, and the x- and y-axes show the log read counts aligning to the site in the respective replicates (top row) or cell types (bottom row). (C) We find that the most proximal genes near cell-type–specific binding sites are differentially expressed between cell types. The cumulative distribution of log expression level changes are shown. Expression is estimated by RNA-seq in units of reads per thousand nucleotides of transcript per million reads aligned (RPKM).