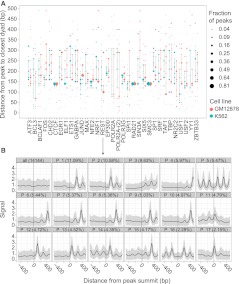
Figure 3.
(A) Distribution of distances between the TF binding site and the closest nucleosome for all TFs assayed in both GM12878 and K562. For each TF in each cell line, we used the median signal of the clusters to compute the distance between the TF binding site and the closest nucleosome positioning peak. The area of each dot is proportional to the fraction of peaks of the TF with the given distance between the binding site and the closest nucleosome dyad. The vertical line extends from the first to the third quartile of distances for each TF. (B) Nucleosome positioning patterns uncovered by CAGT around REST binding sites in K562. (Top, left) A traditional aggregation plot, averaging the signal over all 14,144 REST sites. The rest of the panels show the CAGT clusters in order of prevalence, with the percentage of REST peaks in each shown in the header. Two clusters containing <2% of REST peaks each are omitted from the figure. Note the large diversity of nucleosome positioning shapes, with distances between the binding site and the closest nucleosome positioning peak varying widely from 10 bp (P_17) to 300 bp (P_4).