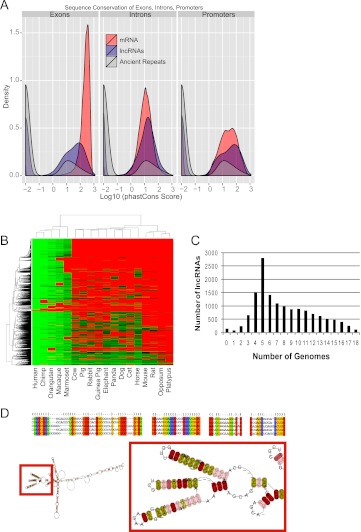
Figure 4.
Evolutionary conservation of lncRNAs. (A) Density plots of phastCons score distributions of protein genes (red curves), lncRNA genes (blue curves), and ancestral repeats (gray curve) for exons (left), introns (middle), and promoters (right). (B) Human lncRNA conservation in mammals: The heatmap summarizes the lncRNA orthologs discovered in 18 other mammalian genomes (see Methods). (Columns) Mammal species. (Rows) Query lncRNAs. The color scheme reflects the level of sequence similarity (percent identity) measured between query and target homologs. (Red) No reliable homolog was detected. (C) The number of orthologs discovered for each lncRNA. LnRNAs with zero orthologs are those that could not be reliably remapped to the human genome at the levels of stringency used in the analysis, due to high repeat content. (D) Example of a multiple sequence alignment of a five-member family. The position containing compensated mutations are labeled by orange columns (correlated) and red columns (correlated Watson-Crick). (Yellow columns) Perfect Watson-Crick matches; (green columns) neutral matches (including G-U pairs); (blue columns) incompatible matches. The putative 2D consensus structure shown is based on the full multiple sequence alignment (RNAaliFold minimum folding energy). (Red box) Details of the 2D structure, with the precise location of the groups of compensated mutations. The colors associated with the residues indicate mutational pattern with respect to the structure as reported by RNAalifold.