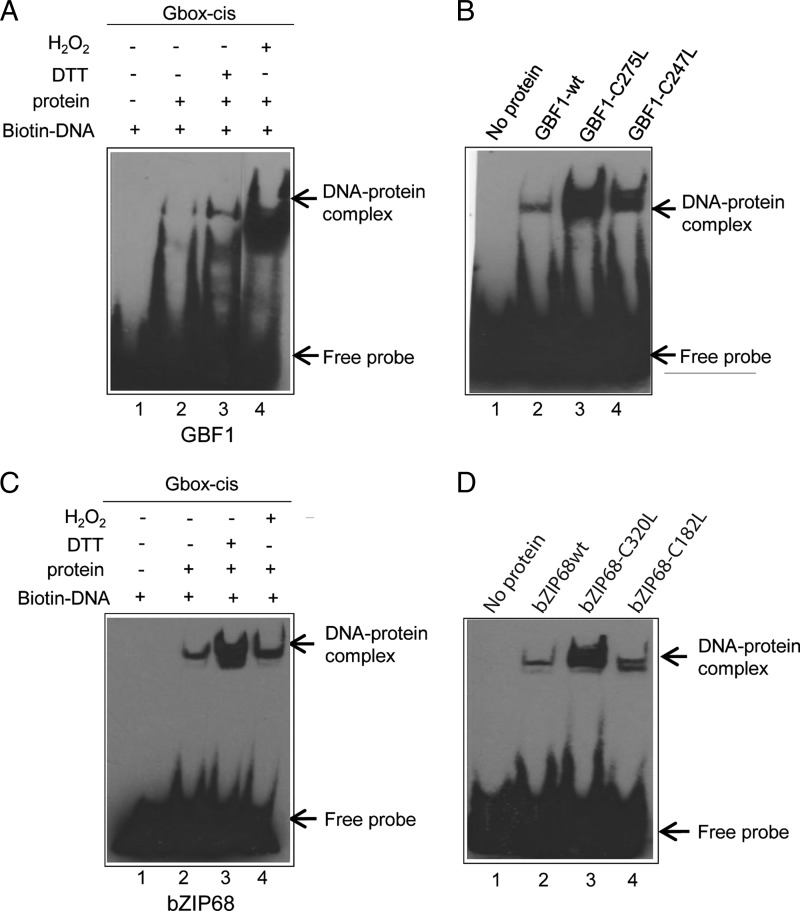
FIGURE 7.
Redox modulation of AtbZIP68 and AtGBF1 binding affinities to the G-box. A, the effect of DTT and H2O2 on DNA binding activity of GBF1 was tested by EMSA. EMSA was performed using biotin-labeled complementary synthetic oligonucleotides representing the G-box element without protein or with GBF1-WT, 10 mm DTT, and 10 mm H2O2. B, binding of GBF1-WT protein and its mutant variants to the G-box element. Binding reactions in the absence of GBF1 protein or with GBF1-WT, GBF1 containing a Cys to Leu mutation at position 275, or GBF1 containing a Cys to Leu mutation at position 247. C, the effect of DTT and H2O2 on DNA binding activity of bZIP68 was analyzed by EMSA. Binding reactions in the absence or presence of bZIP68 protein, 10 mm DTT, and 10 mm H2O2. D, binding of bZIP68-WT protein and its mutant variants to the G-box element. EMSA was performed using the biotin-labeled G-box element without protein or with bZIP68-WT, bZIP86 containing a Cys to Leu mutation at position 320, or bZIP68 containing a Cys to Leu mutation at position 182. Biotin-labeled probes were detected with a chemiluminescent nucleic acid detection module, and positions of free DNA and protein-DNA complexes are indicated by arrows.