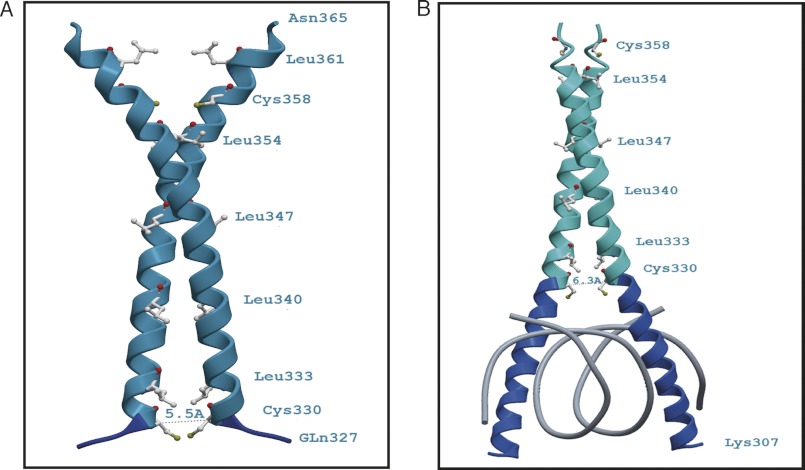
FIGURE 8.
Ribbon diagrams of the theoretical three-dimensional structures (comparative models) of the bZIP domain of AtbZIP16. The figures show the proximity of the cysteine residues at position 330 of the AtbZIP16 sequence without the basic DNA binding region (A) and when bound to DNA (B). A, a model based on amino acids Gln327–Asn365 built onto a DNA-free template (Protein Data Bank code 3HE4_A). The position of Cys330 is indicated, and the distance between the two corresponding Cys330 Cα atoms was determined to be roughly 5.5 Å. At this Cα-Cα distance, the sulfur atoms (Sγ) of the cysteine residues would be sufficiently close to form a disulfide bond. The S-S separation in the model is about 1.5 Å. B, a homology model of residues Lys307–Glu359 built onto the DNA-bound structure of CREB from mouse (Protein Data Bank code 1DH3_A). In this case, the distance between the Cys330 Cα atoms is about 6.3 Å, which would be too far to allow a disulfide bond to form. Dark blue depicts the basic region, and cyan marks the leucine zipper part of AtbZIP16. Amino acids are depicted in gray with the sulfur atoms (Sγ) in yellow and carbonyl oxygen atoms in red.