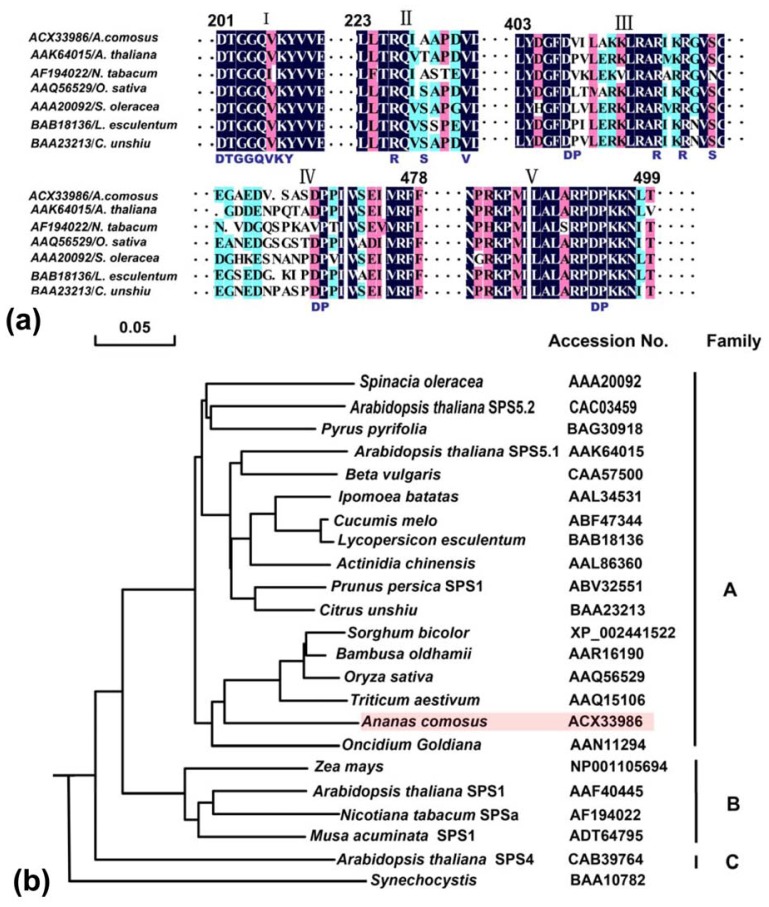
Figure 2.
(a) Conserved regions and phylogenetic analysis of sucrose phosphate synthase (SPS) protein sequences. (I) some considered function domains were uncovered as the putative Fru6P binding site; (II) the 14-3-3 regulated phosphoserine and uridie diphosphate glucose (UDPG) binding domain; (III) the osmotically regulated phosphoserine; (IV, V) various aspartate-proline pairs determined in either spinach or maize or both [19]. Each active site of the various motifs was highlighted in capital letters under the alignment. Numbering of amino acids is according to S. oleracea; (b) the amino acid sequences of the ORFs were initially aligned and tree was calculated by Neighbor-Joing algorithms and rooted with Synechocystis. The GenBank accession numbers and name of the sequences were showed.