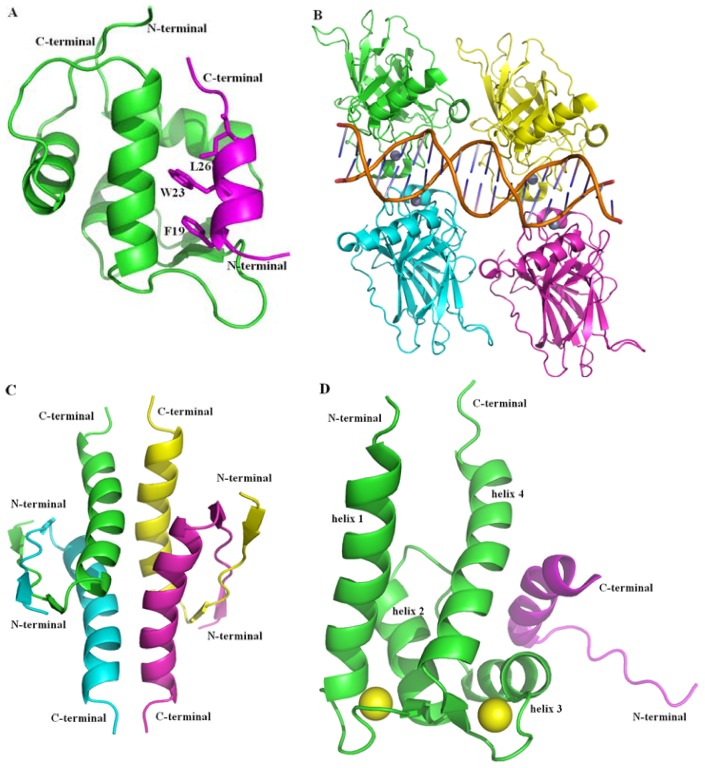
Figure 1.
Structures of p53 protein. (A) The complex of p53 transcriptional activation domain (TAD) fragment bound to MDM2 (PDB 1YCR) [18] is shown in cartoon, p53 TAD fragment (residues 17–29) is shown in magenta and the three most important residues are shown in stick, MDM2 (residues 25–109) is shown in green; (B) The tetramer of the DBD of p53 (PDB 3KMD) [15] is shown in cartoon and the four monomers (residues 92–291) are colored in green, cyan, magenta and yellow, respectively; Zn2+ is shown in sphere and dirtyviolet, and the DNA is shown in stick; (C) The tetramer of oligomerization domain (OD) of p53 (PDB 1PES) [16] is shown in cartoon and the four monomers (residues 325–355) are colored in green, cyan, magenta and yellow, respectively; (D) The complex of p53 C-terminal regulatory domain (CTD) fragment bound to S100 calcium-binding protein B (PDB 1DT7) [19] is shown in cartoon, p53 CTD fragment (residues 377–387) is shown in magenta and yellow, S100B (residues 1–91) is shown in green and cyan and the two Ca2+ are shown in sphere and are colored in, consistent with the S100B protein for the two subunits, respectively. Figures were created with Pymol (http://pymol.org) [20].