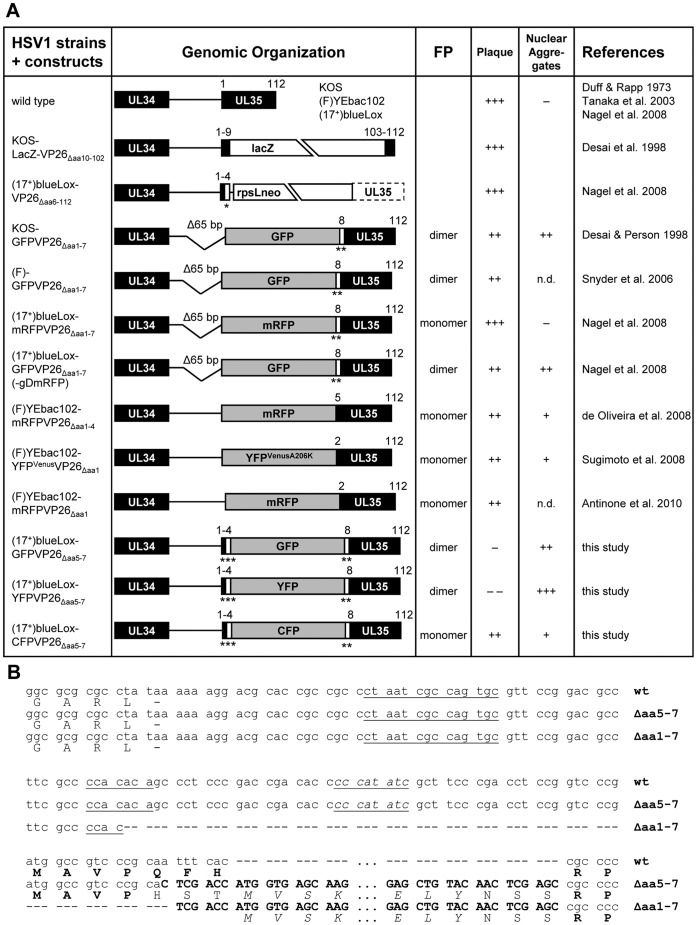
Figure 1. HSV1-VP26 constructs.
(A) 1st column: HSV1 constructs in which the SCP VP26 has been tagged with different fluorescent protein domains. 2nd column: Genomic organization of the UL35 region approximately drawn to scale. The gene UL35 coding for VP26 has been disrupted by replacing it with lacZ or an rpsLneo cassette out of frame. Some constructs lack a 65 bp region upstream of UL35 (D65 bp) including the first seven N-terminal codons of VP26 (Daa1–7), while others lack only four (Δaa1–4) or just one (Daa1) codon. For the present study, the fluorescent protein tag was inserted between VP26 residues 4 and 8 (Daa5–7). Due to the mutagenesis, some strains contain additional linkers (*, AW; **, NSS; ***, HST). 3rd column: Propensity of the fluorescent protein (FP) to dimerize [76]–[80]. 4th column: Ability of the construct to replicate and to form plaques (+++, similar to wild type; ++ attenuated, but robust growth; − strongly attenuated, tiny plaques; −−, single fluorescent cells, no plaques). 5th column: Propensity of the construct to induce nuclear aggregates (+++, large irregular shaped aggregates; ++, large aggregates early after infection or transfection; +, aggregates late in infection; − aggregates in less than 2% of cells even late in infection). 6th column: References. (B) Nucleotide (upper lines) and amino acid (lower lines) sequences of the UL34/UL35 (pUL34/VP26) intergenic region. The 3¢ end of the UL34 ORF until the 5¢ start of the UL35 ORF are shown for wild type HSV-1, the GFPVP26Äaa5–7 (Äaa5–7) and GFPVP26Äaa1–7 (Äaa1–7) mutants. Additional nucleotides inserted during mutagenesis are shown in bold capitals, and the GFP amino acids are shown in italics. Putative Inr late promoter elements are underlined with the element perfectly matching the consensus sequence being underlined and in italics. The original amino acids encoded by UL35 are shown in bold capitals, the inserted GFP residues in italic capitals and the additional linker residues in normal script capitals.