Abstract
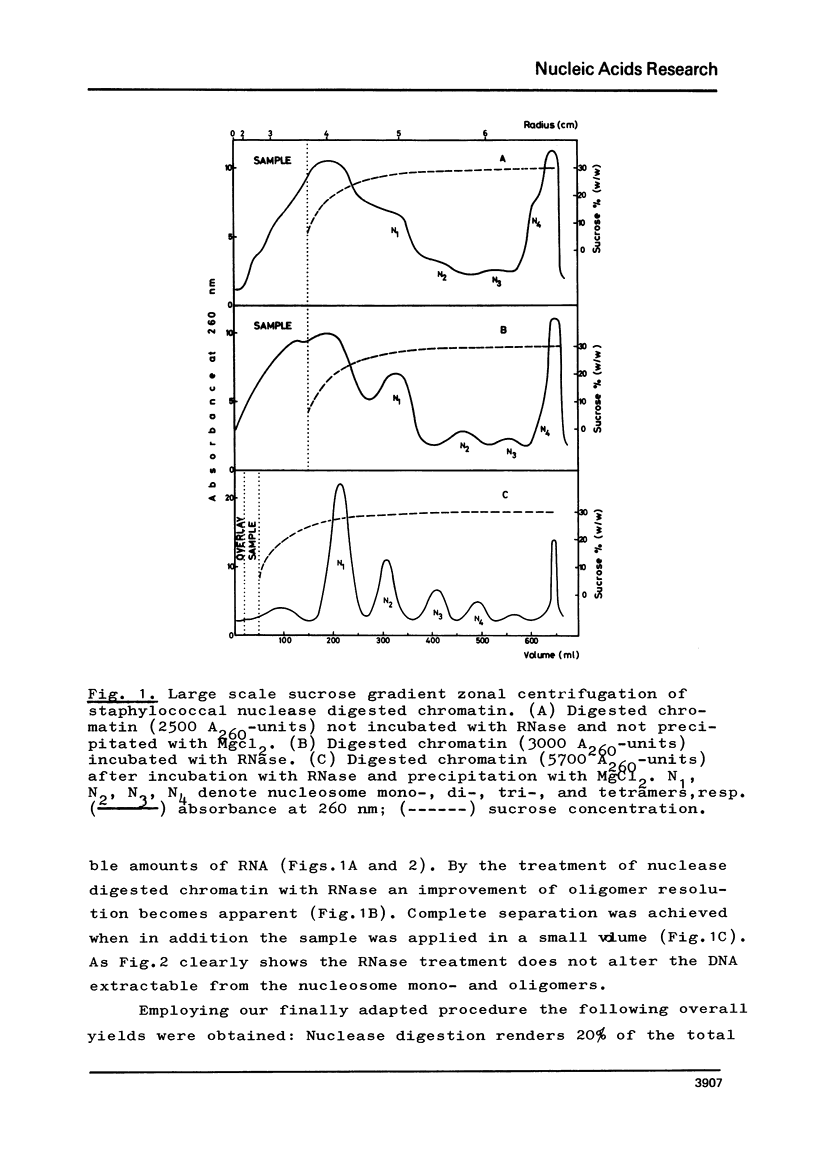
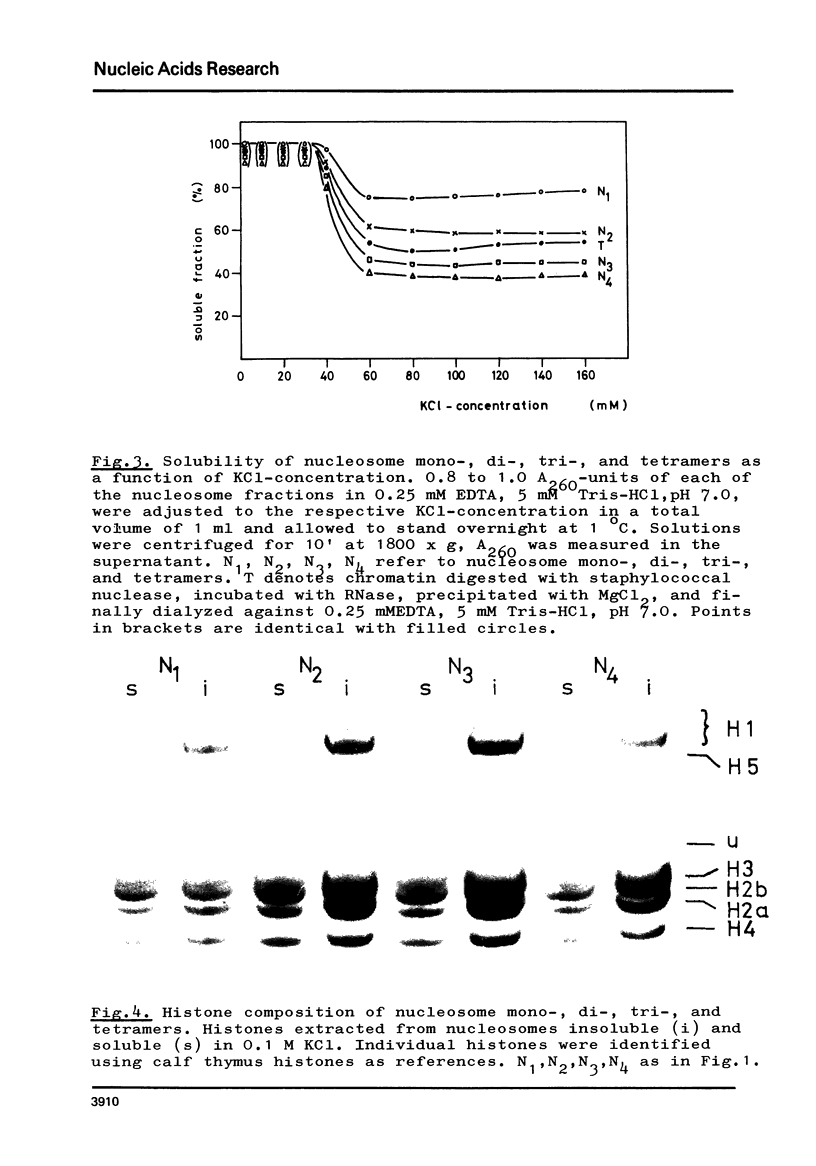
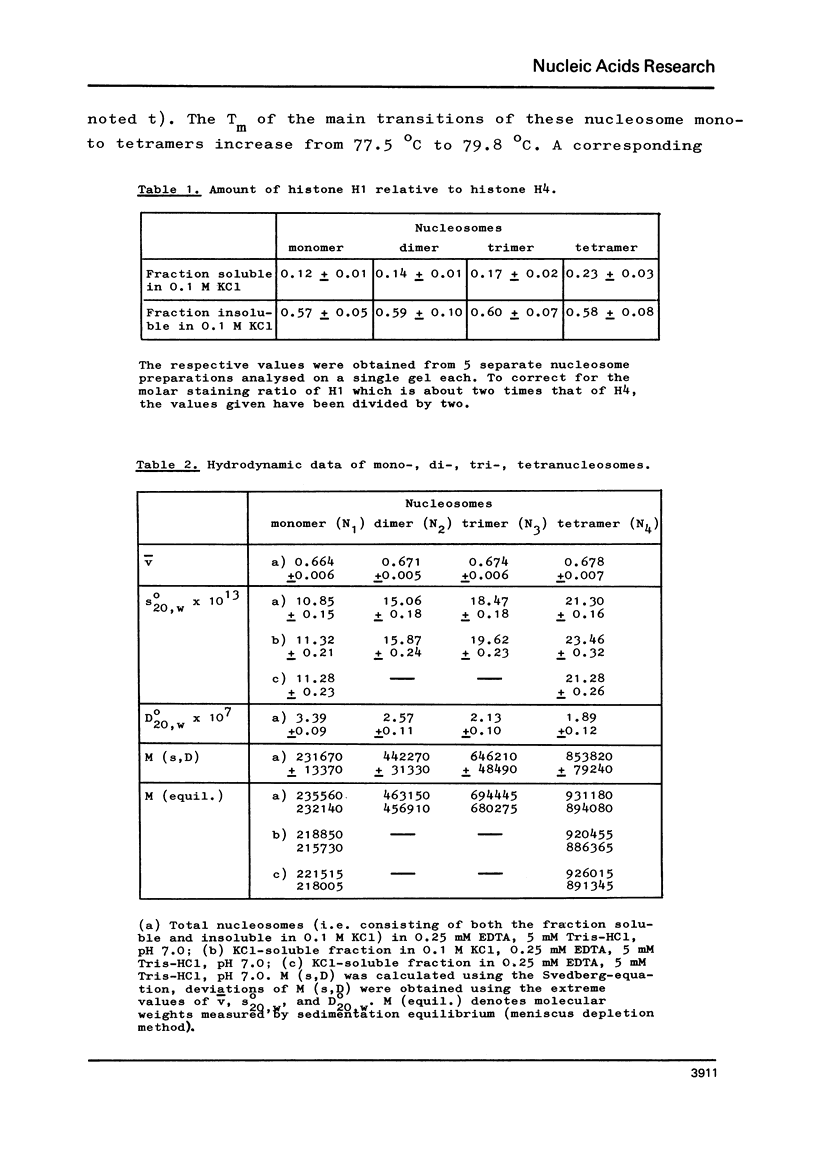
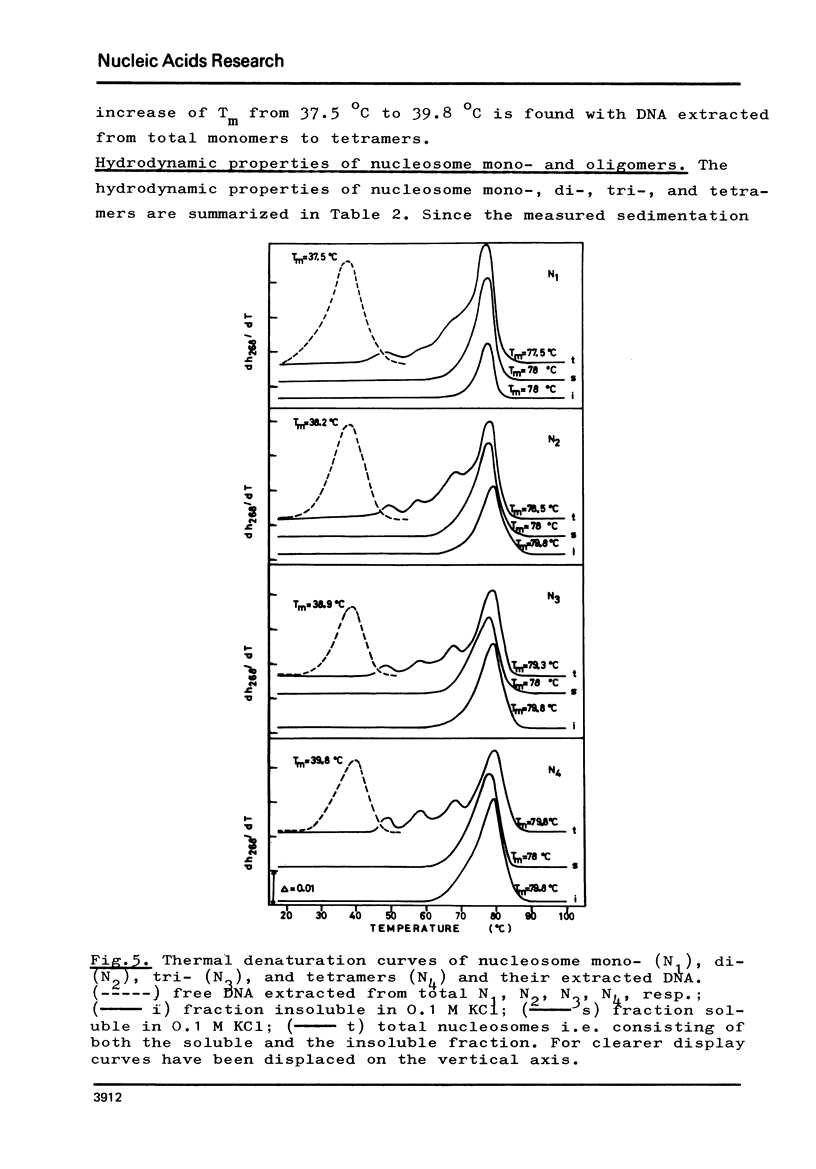
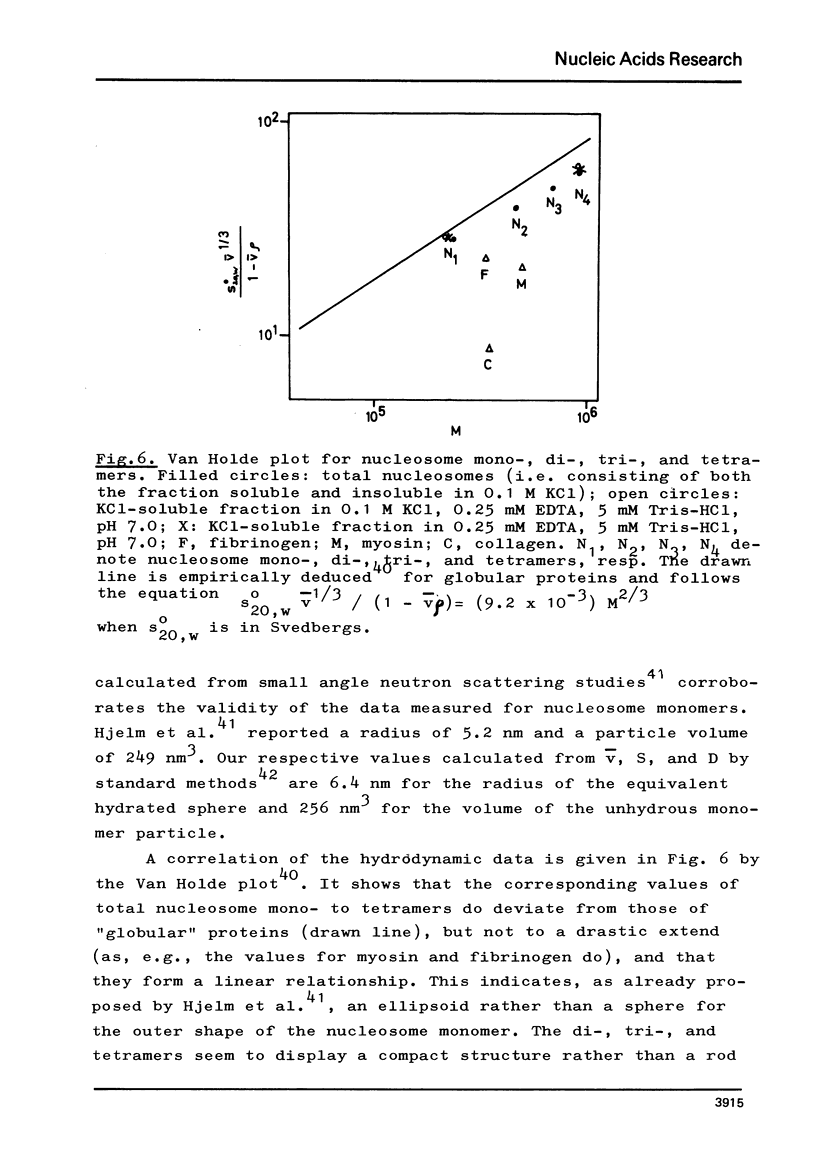
The fractionation of gram quantities of nuclease digested chromatin from chicken embryos into nucleosome mono-, di-, tri-, and tetramers is described in detail. Each of these nucleosomal species contains a fraction soluble in 0-1 M KC1 that decreases with increasing repeat number. Less histone H1 is associated with the nucleosome fractions soluble as compared to the respective fractions precipitated in 0.1 M KC1. Thermal denaturation profiles of the four nucleosomal species are monophasic. The same Tm of 78 degrees C has been determined for the KC1-soluble nucleosomes and for the KC1-insoluble monomer. The Tm of the KC1-insoluble oligomers is 79.8 degrees C. Multiphasic melting curves were recorded for nucleosomal material that was concentrated by lyophilisation or stored at 4 degrees C in 0.25 mM EDTA. Total nucleosome mono-, di-, tri-, and tetramers (consisting of both the fraction soluble and insoluble in 0.1 M KC1) have been analyzed concerning their sedimentation, diffusion, partial specific volume, and molecular weight and compared with the sedimentation and molecular weight data of KC1-soluble nucleosome mono- and tetramers.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakayev V. V., Melnickov A. A., Osicka V. D., Varshausky A. J. Studies on chromatin. II. Isolation and characterization of chromatin subunits. Nucleic Acids Res. 1975 Aug;2(8):1401–1419. doi: 10.1093/nar/2.8.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Ballal N. R., Goldberg D. A., Busch H. Dissociation and reconstitution of chromatin without appreciable degradation of the proteins. Biochem Biophys Res Commun. 1975 Feb 17;62(4):972–982. doi: 10.1016/0006-291x(75)90418-0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Noll M., Kornberg R. D. Electron microscopy of defined lengths of chromatin. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3320–3322. doi: 10.1073/pnas.72.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil W., Igo-Kemenes T., Zachau H. G. Nuclease digestion in between and within nucleosomes. Nucleic Acids Res. 1976 Oct;3(10):2633–2644. doi: 10.1093/nar/3.10.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hjelm R. P., Kneale G. G., Sauau P., Baldwin J. P., Bradbury E. M., Ibel K. Small angle neutron scattering studies of chromatin subunits in solution. Cell. 1977 Jan;10(1):139–151. doi: 10.1016/0092-8674(77)90148-9. [DOI] [PubMed] [Google Scholar]
- Hörz W., Igo-Kemenes T., Pfeiffer W., Zachau H. G. Specific cleavage of chromatin by restriction nucleases. Nucleic Acids Res. 1976 Nov;3(11):3213–3226. doi: 10.1093/nar/3.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Lawrence J. J., Chan D. C., Piette L. H. Conformational state of DNA in chromatin subunits. Circular dichroism, melting, and ethidium bromide binding analysis. Nucleic Acids Res. 1976 Nov;3(11):2879–2893. doi: 10.1093/nar/3.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell T. J. Inhibition of eukaryotic DNA-dependent RNA polymerase release from isolated nuclei and nucleoli by phenylmethylsulfonylfluoride. Arch Biochem Biophys. 1975 Nov;171(1):268–275. doi: 10.1016/0003-9861(75)90032-6. [DOI] [PubMed] [Google Scholar]
- Mandel R., Fasman G. D. Chromatin and nucleosome structure. Nucleic Acids Res. 1976 Aug;3(8):1839–1855. doi: 10.1093/nar/3.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Marshall A. J., Burgoyne L. A. Interpretation of the properties of chromatin extracts from mammalian nuclei. Nucleic Acids Res. 1976 Apr;3(4):1101–1110. doi: 10.1093/nar/3.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Wright E. B., Olins D. E. Chromatin nu bodies: isolation, subfractionation and physical characterization. Nucleic Acids Res. 1976 Dec;3(12):3271–3291. doi: 10.1093/nar/3.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. Sequence analysis of the 3' non-coding regions of rabbit alpha- and beta-globin messenger RNAs. J Mol Biol. 1976 Nov 15;107(4):491–525. doi: 10.1016/s0022-2836(76)80080-0. [DOI] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Preparation and physical characterization of a homogeneous population of monomeric nucleosomes from HeLa cells. Nucleic Acids Res. 1976 Sep;3(9):2255–2266. doi: 10.1093/nar/3.9.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Frado L. L. Thermal denaturation of subchromosomal particles. Biochem Biophys Res Commun. 1975 Sep 2;66(1):403–410. doi: 10.1016/s0006-291x(75)80342-1. [DOI] [PubMed] [Google Scholar]
- Wright E. B., Olins D. E. Histone stoichiometry in chicken erythrocyte nuclei. Biochem Biophys Res Commun. 1975 Apr 7;63(3):642–650. doi: 10.1016/s0006-291x(75)80432-3. [DOI] [PubMed] [Google Scholar]
- YPHANTIS D. A. EQUILIBRIUM ULTRACENTRIFUGATION OF DILUTE SOLUTIONS. Biochemistry. 1964 Mar;3:297–317. doi: 10.1021/bi00891a003. [DOI] [PubMed] [Google Scholar]
- Yu F. L. An improved method for the quantitative isolation of rat liver nuclear RNA polymerases. Biochim Biophys Acta. 1975 Jul 7;395(3):329–336. doi: 10.1016/0005-2787(75)90204-x. [DOI] [PubMed] [Google Scholar]
- van Bruggen E. F., Arnberg A. C., van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. Electron microscopy of chromatin subunit particles. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1365–1370. doi: 10.1016/0006-291x(74)90348-9. [DOI] [PubMed] [Google Scholar]