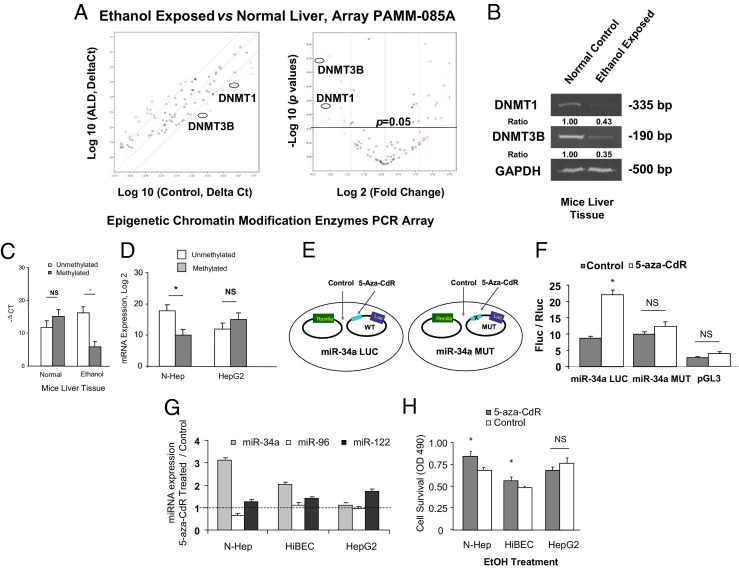
Figure 2.
Epigenetic regulation of miR-34a in ethanol-exposed mouse liver. A: The expression levels of DNMT1 and DNMT3B are down-regulated in ethanol-exposed mouse liver. Relative gene expression profile between ethanol-exposed mouse livers versus control liver tissues is shown. The expression of a panel of diverse epigenetic-associated genes was evaluated by real-time PCR using Mouse Epigenetic Chromatin Modification Enzymes PCR Array (PAMM-085A) from SABiosciences Corporation (Valencia, CA). Gene expression relative to GAPDH was plotted as the Volcano Plots, depicting the relative expression levels (Log10) for selected genes in ALD versus control panels (left). The relative expression levels and P values for each gene in the related samples were also plotted against each other in the scatterplot (right). DNMT1 and DNMT3B are the greatest down-regulated genes among the six epigenetic signaling pathways in ethanol-exposed mouse liver. Data represent mean from three separate experiments. B: RT-PCR confirmed the reduced mRNA expression of DNMT1 and DNMT3B in ethanol-exposed mouse liver relative to controls. Expression of DNMTs was assayed by a standard RT-PCR method in control and ethanol-exposed mouse liver tissues. The PCR amplification was performed for 26 cycles for GAPDH (a housekeeping control), 30 cycles for DNMT1, and 35 cycles for DNMT3B. The ratios shown represent the mean value (relative to control) normalized with GAPDH from four independent experiments. C and D: Methylation-specific PCR analysis of the miR-34a upstream regulatory region was performed in ethanol-exposed mice and normal control liver, as well as ethanol-treated N-Heps and HepG2 cells (100 mmol/L, 7 days). The dark gray bar indicates hypermethylated miR-34a; the white bar indicates unmethylated miR-34a. Long-term alcohol exposure significantly demethylated CpG island enriched 5′-promoter region of miR-34a. miR-34a promoter was also hypomethylated in N-Heps but not in HepG2 cells after ethanol treatment. The results shown represent the mean ± SE from six independent experiments. **P < 0.01 compared with expression in control regions. E and F: Characterization of miR-34a promoter methylation activities by mutation site-specific luciferase assay. Experimental design strategy of miR-34a methylation-specific mutation is presented (E). A 24-bp mutation of the hsa-miR-34a methylation site (106 bp) converted from GCC to TAA was performed using oligonucleotides to the CpG island enriched region. Luciferase reporter constructs containing the sequence of potential methylation site (CpG island enriched region) from the 5′-promoter region of miR-34a inserted downstream of the luciferase gene were generated. miR-34a-LUC (wild type) contains the intact sequence whereas miR-34a-MUT contained the sequence with GCC to TAA nucleotide changes. Reporter constructs were transfected in normal human hepatocytes for 24 hours and then treated with 10 μmol/L 5-Aza-CdR or diluent control for 72 hours (F). The expression of firefly luciferase activity was normalized to that of Renilla luciferase activity for each sample. The increases in relative firefly luciferase activity in the presence of methylation inhibitor in miR-34a-LUC but not in miR-34a-MUT transfected cells indicate the presence of a methylation-modulated target sequence in the 5′-promoter region of miR-34a. The results shown represent the mean ± SE from eight independent experiments. G: N-Heps, HiBECs, and HepG2 cells were treated with 10 μmol/L 5-Aza-CdR for 72 hours. The expression of mature miR-34a, miR-96, and miR-122 was assessed using Taqman real-time PCR assay. 5-Aza-CdR increased miR-34a but not miR-96 expression in normal hepatocytes and cholangiocytes. Data represent mean ± SE from eight separate experiments. H: 5-Aza-CdR treated hepatobiliary cells were subjected to ethanol treatment for 72 hours simultaneously. Cell viability was measured by the MTS assay. Demethylation treatment significantly enhanced cell survival against ethanol in N-Heps and HiBECs but not HepG2 HCC cells. The results shown represent the mean ± SE from four independent experiments. *P < 0.05 relative to controls. NS, no significant difference.