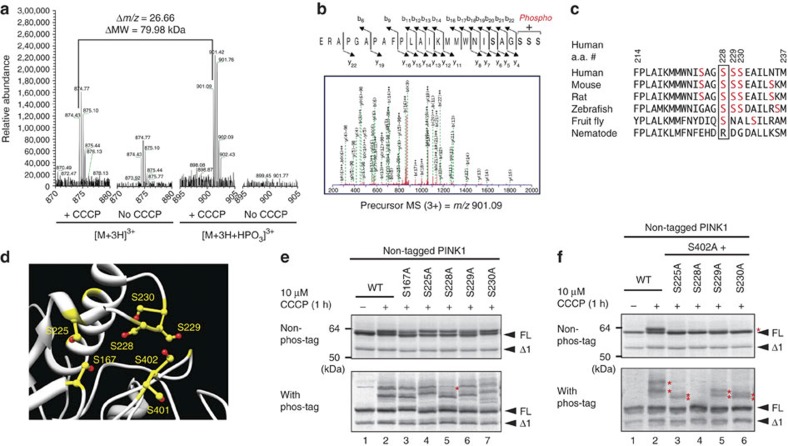
Figure 4. S228 is another autophosphorylation site for PINK1.
(a) Mass spectrometric analysis of the in vivo autophosphorylation site of PINK1. PINK1-GST purified from cells +/−CCCP treatment was subjected to LC-MS/MS analysis with a phosphorylated peptide equivalent to amino acids 206–230 detected only from CCCP-treated cells. (b) The MS/MS data suggested that phosphorylation occurs at Ser228, Ser229 or Ser230. (c) Multiple sequence alignment of PINK1 residues neighboring Ser228 from various organisms. Ser228 (boxed) has been evolutionarily conserved across most species. (d) Structural model (Protein Model Database ID: PM0076345) of PINK1 revealed that the possible phosphorylation sites including S228 and S402 are spatially close to one another. Ser residues are shown in yellow and hydroxyl groups are highlighted in red. (e) The S228A mutation changed the phosphorylation pattern of PINK1. The first band (shown by a red asterisk) in phos-tag PAGE is not observed in cell expressing the S228A mutation. (f) Autophosphorylation-derived signals of PINK1 in cells expressing the S228A/S402A double mutation are completely abolished. Asterisks indicate the phosphorylated form.