Abstract
Regulator of telomere length 1 (RTEL1) is a DNA helicase protein that has been demonstrated to be required for the maintenance of telomere length and genomic stability. It has also been found to be essential for DNA homologous recombination during DNA repairing. Human RTEL1 genomic locus (20q13.3) is frequently amplified in multiple types of human cancers, including hepatocellular carcinoma and gastrointestinal tract tumors, indicating that upregulated RTEL1 activity could be important for tumorigenesis. In this study, we have developed a conditional transgenic mouse model that overexpress mouse Rtel1 in a Cre-excision manner. By crossing with a ubiquitous Cre mouse line, we further demonstrated that these established Rtel1 conditional transgenic mice allow to efficiently and highly express a functional Rtel1 that is able to rescue the embryonic defects of Rtel1 null mouse allele. Furthermore, we demonstrated that more than 70% transgenic mice that widely overexpress Rtel1 developed liver tumors that recapitulate many malignant features of human hepatocellular carcinoma (HCC). Our work not only generated a valuable mouse model for determining the role of RTEL1 in the development of cancers, but also provided the first genetic evidence to support that amplification of RTEL1, as observed in several types of human cancers, is tumorigenic.
Keywords: RTEL1, Mouse transgenic, ES cells, Cre, Overexpression, Hepatocellular carcinoma (HCC)
Introduction
RTEL1 is a newly identified DNA helicase that has been shown to be essential for the maintenance of telomere length and genomic stability (Barber et al. 2008; Ding et al. 2004; Youds et al. 2010). It belongs to the DEAH subfamily of the Superfamily 2 (SF2) helicases and is further classified as a member of DinG, Rad3-related DNA helicase with 5′–3′ directionality (Ding et al. 2004; Fairman-Williams et al. 2010). RTEL1 was identified in an effort to search for the essential factors that govern the telomere length diversity in mammalian species. In mice, Rtel1 gene is located on distal chromosomal 2q region that has previously been shown to be involved in the regulation of mouse species-specific telomere length (Zhu et al. 1998). Mice with Rtel1 deficiency have global telomere loss and multiple genomic instabilities, such as chromosome fusions and breaks (Ding et al. 2004), demonstrating that RTEL1 has an important role in the regulation of telomere length and genomic stability.
More recently, RTEL1 was also found to be required for DNA homologous recombination (HR) (Barber et al. 2008; Youds et al. 2010). HR is one of the major pathways to maintain genomic stability and is involved in the repair of complex DNA damage, including DNA double-strand breaks (DSBs), interstrand crosslinks and DNA gaps. The function of HR in the recovery of stalled and collapsed replication forks is also critical to faithfully duplicate and segregate the genome to daughter cells. Moreover, HR has also been demonstrated to be essential for accurate chromosome segregation during meiosis (see a recent review by Heyer et al. 2010). One critical step in HR is to form a D-loop structure by the invasion of RAD51-single strand DNA filament into homologous DNA template (Heyer et al. 2010; Holthausen et al. 2010; San Filippo et al. 2008). Within D-loops, the invaded DNA strand serves as a primer for DNA synthesis, and the synthesized DNA strand is subsequently displaced and annealed to the other processed DNA end. This SDSA (synthesis-dependent strand annealing) repair pathway leads to yield a non-crossover repair product that can avoid crossover and thus the possibility of deleterious genome rearrangements (Heyer et al. 2010). Therefore, SDSA is considered as the predominant recombinational repair pathway in somatic cells. RTEL1 was demonstrated to promote SDSA by the disruption of D-loops in vitro (Barber et al. 2008). Consistent with this biochemical activity, depletion of RTEL1 in human cells was found to have a fourfold increase in DSB-mediated intrachromosomal recombination (Barber et al. 2008), and C. elegans with Rtel1 mutation showed significantly increased crossover recombination during meiosis (Barber et al. 2008; Youds et al. 2010). These results strongly indicate that RTEL1 has an important function in anti-recombination. Since telomeres can form a large duplex loop (T-loops) that largely resembles the D-loops during HR (Griffith et al. 1999), this function of RTEL1 could also be essential for the replication of telomeres. In the absence of RTEL1, telomeric T-loops could not be efficiently resolved, leading to large telomeric deletions and other telomere dysfunction phenotypes as observed in Rtel1 null or Rtel1 knockdown cells (Ding et al. 2004; Sfeir et al. 2009).
Taken together, several recent studies have established an essential role of RTEL1 DNA helicase in the maintenance of telomere length and genomic stability. Given that telomere dysfunction is dramatically mutagenic and plays an important role in tumor initiation and progression (Maser and DePinho 2002), RTEL1 deficiency is expected to have a tumorigenic function. However, as indicated by human genetic data that RTEL1 genomic locus (20q13.3) is frequently amplified in several human cancers (Bai et al. 2000; Katoh et al. 2006; Muleris et al. 1995; Pitti et al. 1998; Wong et al. 1999), upregulated RTEL1 activity could also be important for tumorigensis. High RTEL1 activity may also induce genomic instability by preventing HR (Uringa et al. 2011), leading to the formation of tumor. As a first approach to determine the role of increased RTEL1 activity in tumorigenesis, in this study, we established a transgenic mouse model that conditionally overexpress mouse Rtel1. We further demonstrated that this established mouse model can serve as a valuable genetic tool to assess the tumorigenic function of upregulated RTEL1 activity in vivo. With this mouse model, we showed for the first time that upregulated RTEL1, as observed in human cancers, is tumorigenic and is able to initiate the formation of hepatocellular carcinoma (HCC) in mice.
Methods
Generation of Rtel1 conditional transgenic mice
The mouse Rtel1 cDNA with a 3′ inserted V5 tag was cloned to a conditional transgenic vector as described previously (Ding et al. 2002). The transgenic vector was verified by restriction enzymes and sequencing.
The transgenic vector was linearized with SacII and then electroporated into R1 mouse embryonic stem (ES) cells. The transfected ES cells were selected with G418 (250 μg/ml). The G418-resistant ES clones were screened by Southern blot analysis using a neo cDNA probe. The genomic DNA was digested with multiple restriction enzymes with unique digestion in the transgenic vector (not in the probe region). Two clones with overall, nonmosaic LacZ expression were used to generate chimeric mice by ES cell⟺diploid embryo aggregation. The transmitting chimeric males were crossed with 129S1 inbred and ICR outbred females to produce hemizygous transgenic offsprings, which were then further bred with EIIa-Cre line (JAX lab, Bar Harbor/Maine) to delete loxP flanked β-geo cassette. The resulted mouse allele, termed pCX-Rtel1-V5, was maintained in a 129S1/C57B6 background. All mouse experiments were performed in accordance with procedures approved by the University of Manitoba Animal Care and Use Committee.
A PCR based genotyping method was applied to genotype pCX-Rtel1-V5 transgenic mice with the primers: forward (5′-TTCGGCTTCTGGCGTGTGACC-3′) and reward (5′-CTCGGAGATGTTGTTGCCAGGC-3′). One pair of primers: forward (5′-AGCCTCTGCTAACCATGTTCA-3′) and reward (5′-TGGGATAGGTTACGTTGGTGT-3′), were used to genotype Rtel1 conditional transgenic mice.
Immunohistochemistry
Mice were transcardially perfused with 4% paraformaldehyde in PBS buffer (pH 7.4). Tissues were dissected, and then post-fixed in the same fixative overnight. After a brief washing with PBS, the fixed tissues were embedded in paraffin. 5 μm Paraffin-embedded mouse tissue sections were pretreated with Retrieval solution (Dako) for 20–30 min at 98°. Pretreated slides were blocked with either mouse IgG blocking reagent (Vector Laboratory) or serum free blocking reagent (Dako), and then incubated with primary antibodies overnight at 4°. Primary antibodies used were rabbit anti-V5 (Abcam, dilution 1:1,000), mouse anti-Ki67 (BD Pharmingen, dilution 1:100), mouse anti-β-catenin (BD Pharmingen, dilution 1:200), and rabbit anti-AFP (Dako, dilution 1:500). After washing with TBS with 0.1% Tween20 buffer, sections were incubated with biotinylated anti-rabbit or anti-mouse antibody for 60 min at room temperature. The sections were washed 3 times with TBS-0.1% Tween20 buffer and incubated with avidin–biotin-peroxidase complex (Vector Laboratory), according to the manufacturer’s protocol. Color was developed through the use of metal 3,3′-diaminobenzidine tetrachloride for 5 min at room temperature.
Western blot analysis
For western blots, mouse liver and lung tissues were homogenized with ice-cold RIPA buffer. 30 μg lysate was separated by 8% SDS-PAGE and transferred to nitrocellulose membrane, which was blocked with 5% de-fatted milk and incubated with mouse anti-V5 antibody (dilution 1:3,000, Invitrogen). Protein was detected using the enhanced chemiluminescence system (Amersham Life Science).
Results and discussion
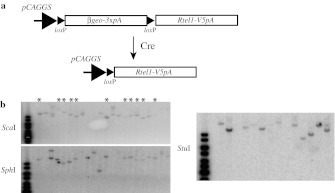
To develop a mouse model that conditionally overexpress mouse Rtel1, we applied a transgenic strategy as described previously (Ding et al. 2002). Briefly, a strong and ubiquitous promoter, pCAGGS (a chicken β-actin promoter with the upstream CMV enhancer) (Niwa et al. 1991), was used to drive the expression of a loxP flanked β-geo cassette which is followed by mouse Rtel1 cDNA. The Rtel1 expression can be turned on only after Cre-mediated excision of loxP flanked β-geo (Ding et al. 2002) (Fig. 1a). To facilitate the detection of Rtel1 protein in transgenic mice, a V5 tag was inserted 3′ downstream of the last coding codon of Rtel1 cDNA. This addition will not affect the cellular function of Rtel1 as demonstrated by the previous studies (Barber et al. 2008; Ding et al. 2004). The transgenic vector (Fig. 1a) was electroporated into R1 ES cells. Clones with single copy insertion were identified by Southern hybridization (Fig. 1b).
Fig. 1.
A transgenic approach for establishing Rtel1 conditional transgenic mice. a Schematic representation of transgenic vector. pCAGGS promoter (the fusion of a chicken β-actin promoter with upstream CMV enhancer) is followed by a loxP flanked β-geo with three SV40 polyadenylation signals (pA) and a mouse Rtel1-V5 transgene. Prior to Cre excision, this transgenic vector expresses only β-geo. The Rtel1-V5 expression is turned on after Cre-mediated excision. b Southern blot analysis of transgene integration in R1 ES cells using a neo cDNA probe. The genomic DNA was digested with restriction enzymes with unique digestion in the transgenic vector (not in the probe region), i.e.. ScaI and SphI. The potential ES clones with a single copy and single site integration of the transgene (labeled by asterisks) was further analyzed by another restriction enzyme with unique digestion, StuI. All these diagnostic digests were consistent with a single copy and single site integration of the transgene
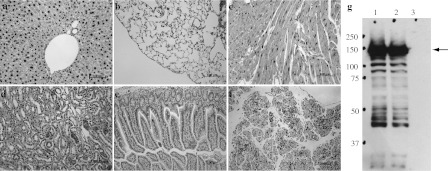
We applied the ES cell⟺diploid aggregation assay to create chimeras from two independently targeted ES clones. Both clones gave germline transmission of transgenic allele. The transmitting chimeric males were crossed with 129S1 inbred and ICR outbred females to produce hemizygous transgenic offspring that appeared normal and were fertile. Without Cre recombinase-mediated excision, these transgenic mice only expressed LacZ detected by X-gal staining (data not shown). To determine Rtel1 expression upon Cre-mediated excision, we crossed Rtel1 conditional transgenic mice with EIIa-Cre transgenic mice (a ubiquitous Cre transgenic line). The resulted mouse allele, termed pCX-Rtel1-V5, was postnatal viable and fertile. We further applied immunohistochemistry (IHC) to determine the expression of Rtel1-V5 protein in pCX-Rtel1-V5 adult mice with anti-V5 antibody. As shown in Fig. 2, strong nuclear-localized Rtel1-V5 protein was found to be widely presented in most tissues, such as liver, lung, heart, kidney, intestine and pancreas. We also applied western blot to demonstrate that a 150 kd full length Rtel1-V5 protein was expressed in pCX-Rtel1-V5 mouse tissues (Fig. 2g). This highly and widely expression of Rtel1-V5 was consistently observed in two independent conditional transgenic lines upon EIIa-Cre-mediated excision, demonstrating that our established Rtel1 conditional transgenic mice allow to efficiently overexpress Rtel1 in vivo.
Fig. 2.
Characterization of Rtel1-V5 protein expression in pCX-Rtel1-V5 mice. a–f IHC with anti-V5 antibody on mouse tissues collected from 2 months old pCX-Rtel1-V5 mice. a liver; b lung; c heart; d kidney; e intestine; f pancreas. g western blot with anti-V5 antibody on mouse tissue lysates harvested from pCX-Rtel1-V5 mice. 1 liver; 2 intestine; 3 liver from wild-type control. Arrow indicates a major detected protein band with 150 Kd that matches with the predicted molecular weight of full length Rtel1-V5 protein
To demonstrate that Rtel1-V5 protein expressed from pCX-Rtel1-V5 transgenic mice is functional, we bred pCX-Rtel1-V5 mice with Rtel1 null mouse allele. We previously showed that Rtel1 −/− mice were embryonic lethal, and died between E9.5 and E10.5 with multiple developmental defects that are characterized by neural tube defect (NTD), bulbous allantois, pericardial effusion and the deficiency of trophoblasts (Ding et al. 2004; Wu et al. 2007). All these phenotypes were not observed in Rtel −/− /pCX-Rtel1-V5 E10.5 embryos. Rtel −/− /pCX-Rtel1-V5 mice were postnatally viable, and showed similar behaviors as other wild-type littermates. This clearly indicates that expressed Rtel1-V5 protein in pCX-Rtel1-V5 transgenic mice has the same cellular function as native Rtel1, which can completely rescue the developmental defects of Rtel1 null mutants.
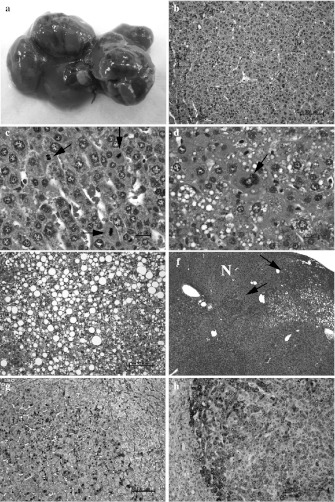
Lastly, we applied pCX-Rtel1-V5 transgenic mice to determine whether upregulated Rtel1 activity could induce the formation of tumors in vivo. For this purpose, we monitored a cohort of 22 mice (12 pCX-Rtel1-V5 mice and 10 wild type age matched littermates) with a normal diet for a one-year period. At 12 months old, 8 out 12 pCX-Rtel1-V5 mice showed weakness and decreased body weight as compared to wild-type littermates. Post-mortem examination revealed these pCX-Rtel1-V5 mice developed numerous discrete nodules throughout the livers (Fig. 3a). Upon histological analyses, these nodules were found to display many pathological characteristics of HCC, which include trabecular growth of multiple layers of tumor cells (Fig. 3b), tumor cells with high mitotic figures (Fig. 3c), cellular pleiomorphism with multinucleated cells (Fig. 3d) or with loss of cytoplasmic staining (Fig. 3e), and the invasion of nodular tumor cells into the surrounding parenchyma (Fig. 3f). Similar to some of human HCC, liver tumors from pCX-Rtel1-V5 mice was also found to have increased β-catenin activity (as reflected by the nuclear accumulation of β-catenin in tumor cells) (Fig. 3g). Some regions of liver tumors from pCX-Rtel1-V5 mice also showed strong positive for α-fetoprotein (AFP) (Fig. 3h), a diagnostic and prognostic marker in human HCC. This liver tumor phenotype was consistently present in two independent pCX-Rtel1-V5 transgenic lines. None of wild-type control mice developed liver tumor at this developmental stage. All these indicate that overexpression of Rtel1 in mouse hepatocytes can induce the formation of liver tumors that recapitulate many malignant features of HCC.
Fig. 3.
Development of liver tumors in pCX-Rtel1-V5 mice. a Representative image of liver from a 12-month old pCX-Rtel1-V5 mouse. Note the widespread tumor nodules throughout the liver. b–f Histological characterization of liver tumors from pCX-Rtel1-V5 mice reveals characteristics of hepatocellular carcinoma. b shows a trabecular growth of multiple layers of tumor cells. c shows increased mitotic figures (arrows indicate) in tumor cells. d shows the multinucleated tumor cells (arrow). e shows the cellular pleiomorphism with loss of cytoplasmic staining. f shows the invasion of nodular tumor cells (arrows indicate) to normal liver region (N). g IHC staining with anti-β-catenin antibody reveals nuclear accumulation of β-catenin in most tumor cells. h IHC staining with anti-AFP antibody. Some regions of tumor cells showed strong positive for AFP
In summary, we have generated a conditional mouse model that highly and efficiently expresses Rtel1 upon Cre-mediated excision. We further confirmed that Rtel1-V5 protein expressed in this mouse model is functional. More importantly, we demonstrated that this established mouse model can serve as a valuable genetic tool to assess the tumorigenic function of upregulated RTEL1 activity in vivo. With this model, we showed that increased expression of Rtel1 in mouse hepatocytes induced the development of HCC (Fig. 3). This finding is consistent with human genetic data that amplification of RTEL1 genomic locus is not only a common genetic alteration in HCC, but also closely associated with its malignancy and progression (Guan et al. 2000; Katoh et al. 2006; Niketeghad et al. 2001; Taniguchi et al. 2010; Wong et al. 1999). Therefore, our data indicate that upregulated RTEL1 activity could be a new genetic factor involved in liver carcinogenesis. Future mechanistic studies are required to provide insight into the nature of the genomic instability in these tumors.
Given that RTEL1 gene is also frequently amplified in other human cancers, such as gastrointestinal tract tumors and breast cancers (Bai et al. 2000; Muleris et al. 1995; Pitti et al. 1998), our established Rtel1 transgenic mouse model, which allows to overexpress Rtel1 in a variety of cells or tissues, can also be applied to determine the role of RTEL1 in the development of these human cancers.
Acknowledgments
This work is supported by the Canada Research Chair program and an operating grant from Manitoba Institute of Child health (MICH). S.S. and Z.N. are supported by the fellowships from NSERC (Natural Sciences and Engineering Research Council of Canada) (to S.S.) and the Manitoba Health Research Council (MHRC) (to Z.N.). H.D. is the holder of Canada Research Chair.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Bai C, Connolly B, Metzker ML, Hilliard CA, Liu X, Sandig V, Soderman A, Galloway SM, Liu Q, Austin CP, Caskey CT. Overexpression of M68/DcR3 in human gastrointestinal tract tumors independent of gene amplification and its location in a four-gene cluster. Proc Natl Acad Sci USA. 2000;97:1230–1235. doi: 10.1073/pnas.97.3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, Tissenbaum H, West SC, Rose AM, Boulton SJ. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Wu X, Nagy A. Mice with cre recombinase activatable PDGF-C expression. Genesis. 2002;32:181–183. doi: 10.1002/gene.10065. [DOI] [PubMed] [Google Scholar]
- Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, Tam PP, Nagy A, Lansdorp PM. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/S0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Guan XY, Fang Y, Sham JS, Kwong DL, Zhang Y, Liang Q, Li H, Zhou H, Trent JM. Recurrent chromosome alterations in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 2000;29:110–116. doi: 10.1002/1098-2264(2000)9999:9999<::AID-GCC1022>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair (Amst) 2010;9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Katoh H, Shibata T, Kokubu A, Ojima H, Fukayama M, Kanai Y, Hirohashi S. Epigenetic instability and chromosomal instability in hepatocellular carcinoma. Am J Pathol. 2006;168:1375–1384. doi: 10.2353/ajpath.2006.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- Muleris M, Almeida A, Gerbault-Seureau M, Malfoy B, Dutrillaux B. Identification of amplified DNA sequences in breast cancer and their organization within homogeneously staining regions. Genes Chromosomes Cancer. 1995;14:155–163. doi: 10.1002/gcc.2870140302. [DOI] [PubMed] [Google Scholar]
- Niketeghad F, Decker HJ, Caselmann WH, Lund P, Geissler F, Dienes HP, Schirmacher P. Frequent genomic imbalances suggest commonly altered tumour genes in human hepatocarcinogenesis. Br J Cancer. 2001;85:697–704. doi: 10.1054/bjoc.2001.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- Pitti RM, Marsters SA, Lawrence DA, Roy M, Kischkel FC, Dowd P, Huang A, Donahue CJ, Sherwood SW, Baldwin DT, Godowski PJ, Wood WI, Gurney AL, Hillan KJ, Cohen RL, Goddard AD, Botstein D, Ashkenazi A. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396:699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Yamada T, Sasaki Y, Kato K. Genetic and epigenetic characteristics of human multiple hepatocellular carcinoma. BMC Cancer. 2010;10:530. doi: 10.1186/1471-2407-10-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uringa EJ, Youds JL, Lisaingo K, Lansdorp PM, Boulton SJ. RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 2011;39:1647–1655. doi: 10.1093/nar/gkq1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, Lai P, Lee SW, Fan S, Pang E, Liew CT, Sheng Z, Lau JW, Johnson PJ. Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis: relationship to disease stage, tumor size, and cirrhosis. Am J Pathol. 1999;154:37–43. doi: 10.1016/S0002-9440(10)65248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Sandhu S, Ding H. Establishment of conditional knockout alleles for the gene encoding the regulator of telomere length (RTEL) Genesis. 2007;45:788–792. doi: 10.1002/dvg.20359. [DOI] [PubMed] [Google Scholar]
- Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, OerN NJ, Rose AM, West SC, Meyer BJ, Boulton SJ. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Hathcock KS, Hande P, Lansdorp PM, Seldin MF, Hodes RJ. Telomere length regulation in mice is linked to a novel chromosome locus. Proc Natl Acad Sci USA. 1998;95:8648–8653. doi: 10.1073/pnas.95.15.8648. [DOI] [PMC free article] [PubMed] [Google Scholar]