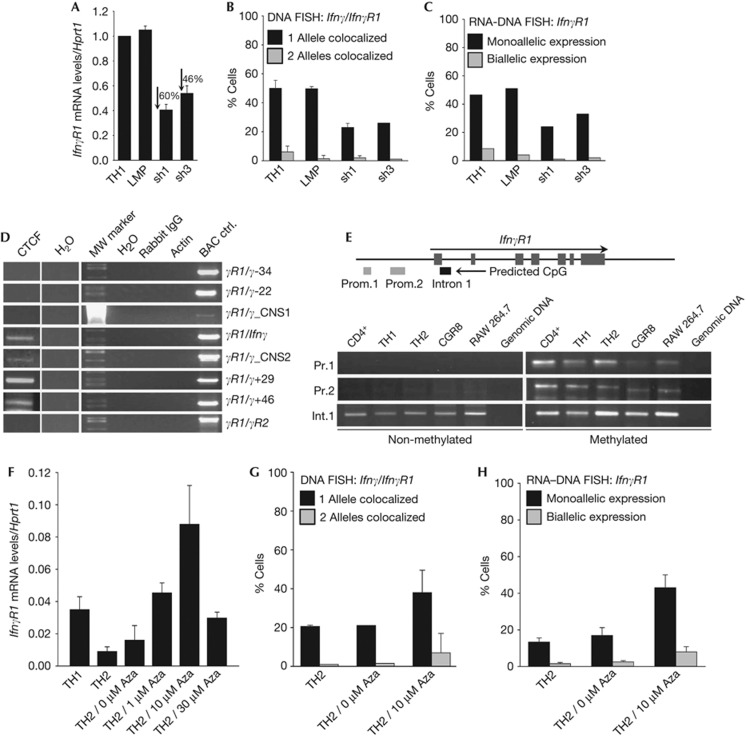
Figure 5.
CTCF mediates the Ifnγ–IfnγR1 intrachromosomal interactions in CD4+ T cells. (A) Quantitative RT–PCR for the IfnγR1 mRNA levels in TH1 cells before and after retroviral transduction of short-hairpin RNAs targeting CTCF. Results are the mean±s.e.m. from triplicate samples from one representative of three independent experiments. (B) DNA-FISH for the Ifnγ-IfnγR1 loci in cells as in (A). Bars are mean values for the percentage of cells with s.d. from six independent experiments for TH1 cells, three experiments for TH1/LMP cells and three experiments for TH1/sh1-CTCF cells. One representative of two conducted is shown for TH1/sh3-CTCF cells. A total of 1178 cells have been scored in total. (C) RNA–DNA FISH for the IfnγR1 gene in cells as in (B). Bars indicate the percentage of cells from one representative experiment of two conducted for each treatment. A total of 568 cells have been scored in total. (D) ChIP-loop analysis using a CTCF and control antibodies and primers designed to detect physical interactions between fragments of the Ifnγ locus and the promoter region of the IfnγR1 gene. (E) PCR analysis of bisulphate-treated genomic DNA [19]. (F) Quantitative RT–PCR for the IfnγR1 mRNA levels in TH2 cells treated with increased concentrations of the DNA demethylating agent 5-Aza-2′deoxycytidine. Results are the mean±s.e.m. from triplicate samples from one representative of three independent experiments. (G) DNA-FISH for the Ifnγ-IfnγR1 loci in TH2 cells, before and after treatment with 5-Aza-2′deoxycytidine. Bars are mean values for the percentage of cells with s.d. from seven independent experiments for TH2 cells and three experiments for TH2 10 μM Aza. The results for TH2 0 μM Aza-treated cells are from one representative experiment of two conducted for this treatment. A total of 1248 cells have been scored in total. (H) RNA–DNA FISH for the IfnγR1 gene in TH2 cells as (G). ChIP, chromatin immunoprecipitation; DNA-FISH, DNA fluorescence in situ hybridization; LMP, empty retroviral vector control; mRNA, messenger RNA; RT–PCR, reverse transcriptase PCR.