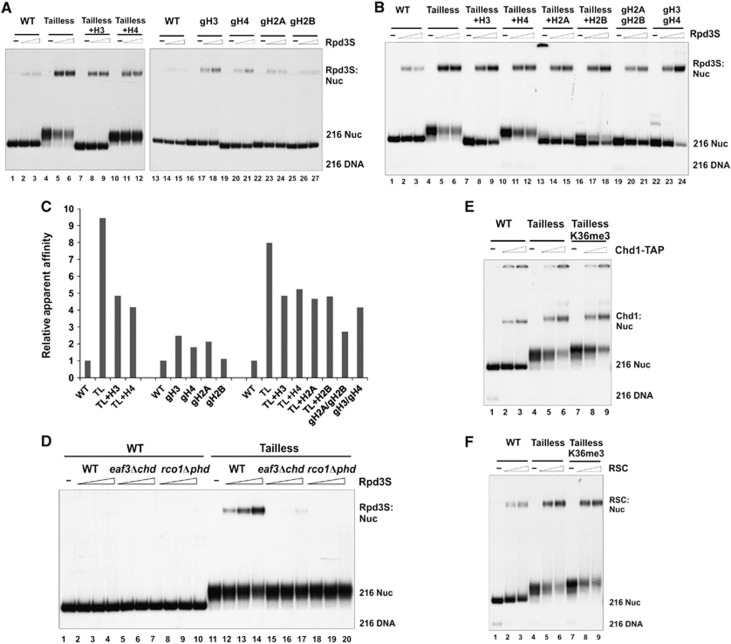
Figure 2.
Rpd3S can contact nucleosomes in a K36me-independent manner. (A–C) The binding of Rpd3S is inversely proportional to the number of histone tails within nucleosomes. (A, B) Gel-mobility shift assays using various forms of WT or tailless nucleosomes. The nomenclature of tail-truncated nucleosomes follows the general rules listed in Figure 1. Tailless (or TL): gH3/gH4/gH2A/gH2B; Tailless+H3: H3/gH4/gH2A/gH2B; Tailless+H4: gH3/H4/gH2A/gH2B; Tailless+H2A: gH3/gH4/H2A/gH2B; Tailless+H2B: gH3/gH4/gH2A/H2B; gH2A/gH2B: H3/H4/gH2A/gH2B; gH3/gH4: gH3/gH4/H2A/H2B; gH3: gH3/H4/H2A/H2B; gH4: H3/gH4/H2A/H2B; gH2A: H3/H4/gH2A/H2B; gH2B: H3/H4/H2A/gH2B. (C) Relative apparent affinities of Rpd3S to each tailless nucleosome. The intensity of gel-shift bands from each nucleosome were quantified using the USI imaging processing software. The ratio between mutant and WT nucleosomes (on the same gel) was defined as relative apparent affinity. (D) The binding of Rpd3S to tailless nucleosome requires PHDRco1 and CHDEaf3. (E, F) Preferential binding to tailless nucleosomes is common among chromatin-associated factors. Two chromatin remodelers (Chd1-TAP (E) and RSC (F)) were tested for their ability to bind to tailless nucleosomes. Both complexes showed increased affinity to tailless nucleosomes, although neither could distinguish between H3K36 methylated and unmodified nucleosomes.