Abstract
Two cationic polyfluorene derivatives, quaternary amine 1 and guanidine 2 sheathed systems, were prepared as potential carriers to mediate import of proteins into cells without requiring covalent attachment to the protein. Neither polymer showed significant cytotoxicities (IC50 100 μM) when exposed to Clone 9 rat liver cells. Both polymers were shown to mediate import of a series of four proteins chosen because they have different pI values, sizes, and variable organic fluor attachments. Once inside the cells, the quaternary amine system 1 released more of its cargo into regions outside the lysosomes. In one exploratory experiment, pyrenebutyrate was shown to accelerate import of a protein system by polymer 1.
Introduction
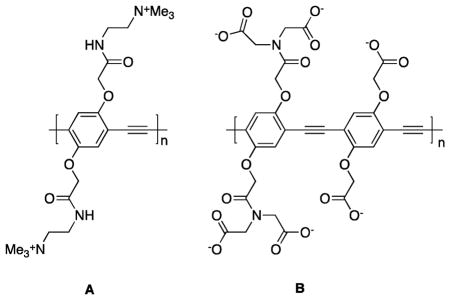
Interactions of polymers with cells depend on a complex interplay of chemical and physical properties. For instance, the cationic poly(p-phenyleneethynylene) material A is imported into punctate endosomal structures within NIH 3T3 fibroblast cells, whereas the corresponding anionic polymer binds to fibronectin fibrils on the surface of those cells.1
There is much to be learned about polyfluorene derivatives interacting with cells, and there is considerable potential for exploiting this knowledge. Fluorene monomers can be easily derivatized to incorporate charges, impart water solubilities, and be conjugated to biomolecules.2 Moreover, polyfluorenes are brilliant and conspicuous under fluorescence microscopes due to their high extinction coefficients and fluorescence quantum yields. Consequently, polyfluorene (or dendric fluorene) derivatives have been tested for cell permeation and staining of organelles,3 transfection of DNA plasmids,4–6 and import then release of cytotoxic materials.7
We are particularly interested in non-covalent import of proteins into cells for imaging intracellular biochemical events. Covalently bound carriers based on HIV-1 Tat and Drosophila Antennapedia homeodomain proteins8,9 are common, but agents that can simply be added to protein cargoes to bring about import are much rarer. Some of the first non-covalent protein import systems include Chariot®,10 the cationic pyridinium amphiphile and a helper lipid, SAINT-PhD,11 BPQ24 BioPORTER® QuikEase™ (a protein delivery kit of unspecified composition),12 the peptide K16SP,13 a cationic polymer containing cholesterol,14 and the somewhat cytotoxic polyethyleneimine (PEI).15,16 None of these systems are perfect, and many import proteins into living cells but deposit them in endosomes or lysosomes where they are degraded. Consequently, there is considerable interest in finding simpler systems that release protein cargoes into the cytoplasm of living cells. For instance, the oligomers of arginine, R8 and R16 have been reported to facilitate significant cellular uptake, while R4 gave relatively little internalization.17,18 However, we showed that these systems tend to result in endosome-trapped proteins, unless the import was carried out at reduced temperatures (4 °C).19,20 In any case, oligomers of arginine are not particularly easy to make.21 Meanwhile others have observed that delivery of proteins into cells can be influenced by lipophilic counter-ions like pyrenebutyrate.22,23
In the research described here, we investigated how the simple, and readily accessible, cationic polyfluorenes might function with respect to import of proteins into living cells. Proteins of different sizes and pI values were used, and they were either labeled with small molecule fluors (FITC, Texas Red®, Alexa Fluor® 488) or chosen because they are intrinsically fluorescent (green fluorescent protein, GFP). The objectives of this study were to establish where the protein and polymer are delivered to inside cells, if indeed they were imported at all.
Results and discussion
Design and synthesis of potential polyfluorene carriers
Amino functionalized polyfluorenes have been made before24 but never exploited as protein delivery systems. Our early studies showed that only the quaternary ammonium system 1 was water soluble; corresponding primary amine, secondary N-methylamine, and tertiary N,N-dimethyl amines polymers were all surprisingly insoluble in aqueous media, even at reduced pHs. However, the quaternary salt 1, very similar to a known material,25 gave a clear solution in water even at neutral pH. Consequently we made a batch of this material via a modification of the literature procedure25 that involved similar steps but in a different order (see ESI†).
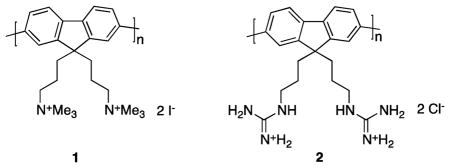
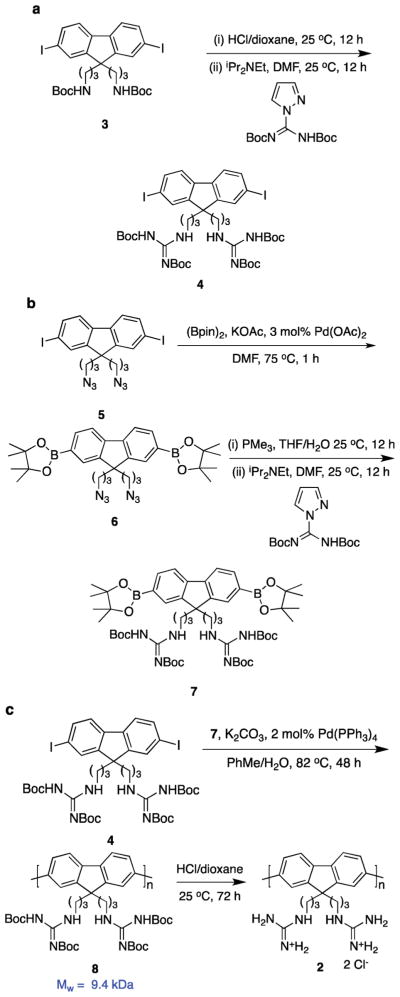
Material 1 is a polyamine derivative whereas the oligoarginines in HIV-TAT mimics contain multiple guanidine functionalities. Consequently, we also prepared the guanidine-functionalized polyfluorene 2. Synthesis of one monomer for the preparation of polymer 2 was achieved via modification of the protected diamine 3 into the masked guanidine 4 (Scheme 1). The complementary diboronate 7 was prepared via the azide intermediates 5 and 6. Finally, Suzuki coupling of monomers 4 and 7, then deprotection afforded the target material 2.
Scheme 1.
Synthesis of polymer 2.
Polymers 1 and 2 had the weight average molecular weight (Mw) of 8200 (Poly Dispersity Index, PDI, 1.21) and 9400 (1.25), respectively, from gel permeation chromatography (GPC). Dynamic light scattering experiments (DLS) indicated polymer 1 in water aggregated to form particles that had average diameters of 140 nm. Polymer 2 was insoluble in water, but it appeared to dissolve in methanol while DLS indicated it had in fact aggregated to form particles with an average diameter of 45 nm. Both polymers exhibited a strong UV absorbance centered on 380 nm, and they fluoresced around 415 nm in water with quantum yields of 0.58 (1) and 0.42 (2).
We were particularly interested in the cytotoxicities of these nanoparticles, so they were tested using Clone 9 rat liver cells in an MTT assay. No significant cytotoxicity was observed for either polymer (IC50 100 μM for both; polymer 2 was delivered as a suspension in buffer). Based on these data it appeared that 1 and 2 were suitable for testing with regards to their abilities to act as carriers for protein import.
Delivery at 37 °C: uptake into punctate vesicular structures
Four proteins were selected for uptake experiments (Table 1). These were cationic (avidin) or anionic (GFP, β-galactosidase, streptavidin), with molecular masses between 26.9 and 540 kDa, and labeled with anionic dye (FITC, Alexa Fluor® 488), cationic dye (Texas Red® ), or no dye at all (GFP). Two cell lines, Clone 9 and Chinese hamster ovary (CHO), were used in the import studies.
Table 1.
Proteins for cellular uptake studies
| Protein | Molecular Weight (kDa) | Amino Acids | pI (unlabelled protein) |
|---|---|---|---|
| Avidin-FITC | 66–68 | 512 | 10–10.5 |
| GFP | 26.9 | 238 | 5.3 |
| Streptavidin-Texas Red® | 52.8 | 4 × 159 | 4.5 |
| β-galactosidase-Alexa Fluor® 488 | 540 | 1171 | 4.8 |
Polymeric carrier-to-protein cargo ratios were optimized so that the signals observed in the cells were visible yet not saturated. A 1 : 1 ratio (mol : mol throughout) was thus found to be optimal for the two proteins of intermediate size. Conversely, less carrier was used for the smallest protein (GFP; 1 : 3 carrier : cargo), and more was necessary for the largest (β-galactosidase; 25 : 1.0 carrier : cargo). In each case, images for the uptake were recorded after 12 h of incubation, then checked after another 24 h. However, the signal for GFP was almost completely lost after 24 h indicating degradation, but the polymer fluorescence was still observed.
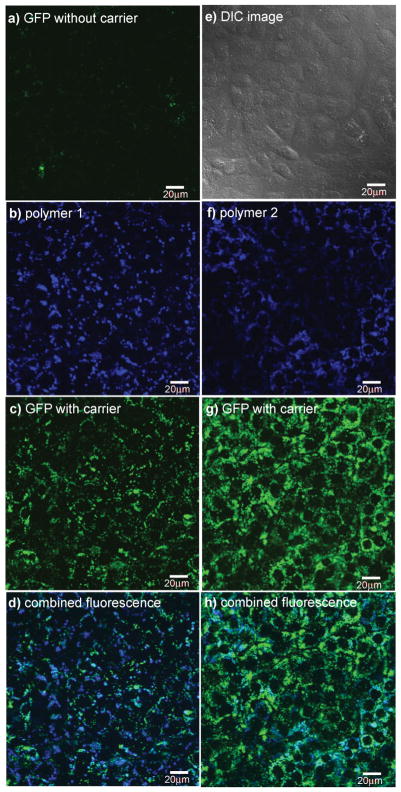
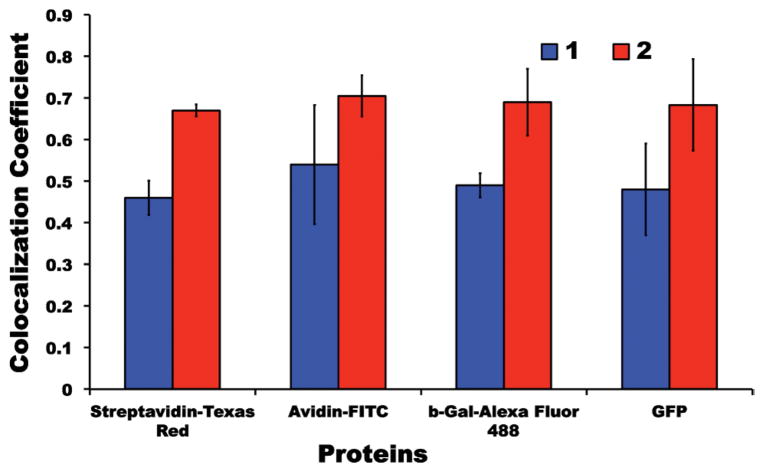
Fig. 1 gives illustrative data for the cellular uptake experiments. This graphic features GFP, but similar images were obtained for all the proteins in both cell lines (see ESI†). Without carrier, little or no visible GFP was imported into the cells (Fig. 1a). A polyfluorene based polymer similar to 1 (different alkyl groups, and another monomer included) has been reported to be imported into cells.26 Consequently, we were not surprised that polymer 1 alone permeated into the cells (data not shown). Polymer 1 and GFP incubated with the cells for 12 h resulted in import of both fluorescent molecules (b and c). Fig. 1d shows the extent of fluorescence overlap from the polymer and protein; this may be expressed in terms of a “colocalization coefficient” deduced using laser scanning microscopy (LSM) software, with larger coefficients corresponding to greater overlap. In this particular case, i.e. for GFP and polymer 1, the coefficient was 0.48. After a further 24 h the signals were weaker, but no diffuse fluorescence was observed. Polymer 2 under the same conditions gave noticeably more uptake of GFP in experiments for which the polymer signal inside the cells was qualitatively similar to the those with GFP/polymer 1. However, more of the GFP colocalized with 2 (colocalization coefficient 0.68) than with polymer 1. Images for import of the other proteins are shown in the ESI,† and Fig. 2 summarizes the colocalization coefficients. Overall, more protein was released from the quaternary amine polymer 1 than it was from the guanidine 2.
Fig. 1.
Delivery of GFP into Clone 9 cells at 37 °C mediated by polymers 1 and 2; a represents no carrier used, and e is a differential interference contrast (DIC) image. Data for polymer 1: b channel showing fluorescence by polymer 1; c channel showing GFP import mediated by 1; d combined fluorescence for GFP and 1 inside the cells. Data for polymer 2: f channel showing fluorescence by polymer 2; g channel showing GFP import mediated by 2; h combined fluorescence for GFP and 2 inside the cells. Throughout, the Clone 9 cells were incubated with the carrier (1.0 μM) and GFP (3.0 μM) at 37 °C for 12 h; the cells were then washed 3×with PBS buffer and 3×with heparin and analysed by fluorescence microscopy. Scale bar is 20 μm.
Fig. 2.
Colocalization coefficients for polymers 1 (blue) and 2 (red) for the four featured proteins in Clone 9 cells.
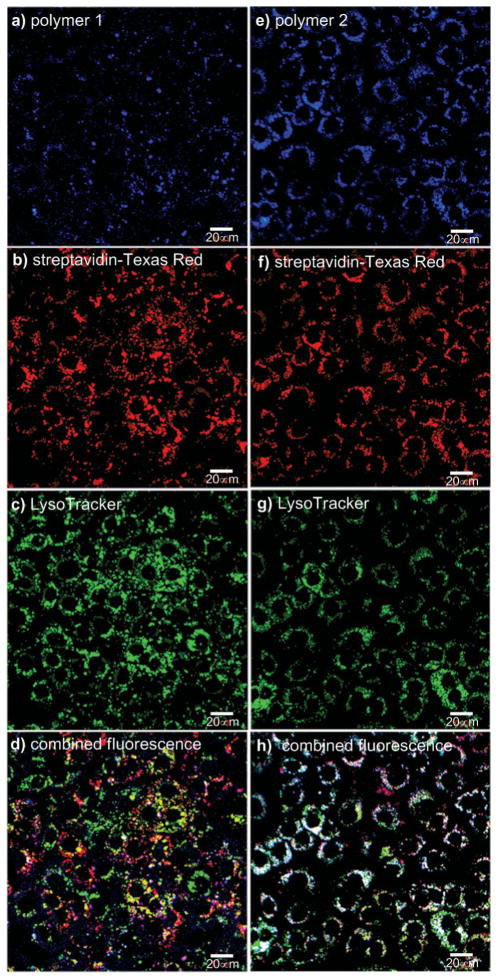
Attempts to establish the location of the protein and polymer were made for each of the proteins imported. Fig. 3 shows the data obtained to track the quaternary amine coated polymer 1, streptavidin-Texas Red®, and LysoTracker® Green. Three dyes are involved in these experiments and all of them show up distinctly (a shows channel to detect polyfluorene; b streptavidin-Texas Red® ; c LysoTracker® Green). Three colocalization coefficients are significant in these experiments: LysoTracker® Green/polymer 1, LysoTracker® Green/streptavidin-Texas Red®, and polymer 1/streptavidin-Texas Red®. These coefficients were 0.35, 0.56, and 0.46, respectively. This data indicates most of the polymer has escaped from the lysosomes, and less than half of the fluorescent protein escaped. Correlations between the polymer 1 and streptavidin-Texas Red® do not distinguish if they are together in the lysosomes, elsewhere, or both. However, Fig. 3d and 3h show areas where all three fluorescent entities are colocalized in white. Quantitative analysis of this situation is difficult and somewhat unnecessary because it is evident that for the quaternary amine polymer 1 regions with distinct blue, red, and green signals predominate; conversely, white areas indicating colocalization of all three labels are prevalent for the guanidine-sheathed polymer 2.
Fig. 3.
Delivery of streptavidin-Texas Red® into Clone 9 cells at 37 °C mediated by 1 and 2. Data for transport using polymer 1: (a) polymer 1 channel; (b) streptavidin-Texas Red® channel; (c) LysoTracker® Green; (d) colocalization shows mostly distinct blue, red, and green areas. Data for transport using polymer 2: (e) polymer 2 channel; (f) streptavidin-Texas Red® channel; (g) LysoTracker® Green; (h) colocalization shows mainly white areas where all three labels coexist. Throughout, the carrier (1.0 μM), streptavidin-Texas Red® (1.0 μM), and the Clone 9 cells were incubated at 37 °C for 15 h; the cells were then washed 3× with PBS buffer and 3× with heparin and analysed via fluorescence microscopy. Scale bar is 20 μm.
Colocalization experiments that parallel those above were attempted with markers for other organelles. Specifically, streptavidin-Texas Red® was tested with compounds that are thought to localize in endosomes, and in the endoplasmic recticulum (ER). However, no significant colocalization was observed.
Similar series of experiments were attempted for the three other proteins. However, the labels involved (GFP, FITC, and Alexa Fluor® ) are all green, so LysoTracker® Green could not be used. LysoTracker® Red was tested, but crosstalk between the red and green channels precluded accurate analyses. Other labels could have been tested, but the chances of success were only moderate, and we made the decision that the scientific gains made by determining the degree of colocalization for these three proteins were not worth the costs and effort involved.
In previous studies, we had found that (Arg)8 mediated import of some of the same protein cargoes into lysosomes at 37 °C, but into the cytosol at 4 °C.19 Experiments to test import with polymers 1 and 2 at 4 °C showed both polymers impregnated into the cell wall membrane, and none of the four proteins were imported.
Futaki and co-workers have studied the influence of the lipophilic, fluorescent counter ion, pyrenebutyrate, in import mediated by oligoarginine-based cell penetrating peptides.22,23,27 Here import of bovine serum albumin-Texas Red® (BSA, MW = 66 kDa, 538 amino acids, pI = 4.7) mediated by polymer 1 was briefly tested in the presence of pyrenebutyrate. This counterion dramatically increased the rate of transport into the cells at 37 °C so that the protein was conspicuous after only 5 min incubation. However, most of the protein colocalized with LysoTracker® Green and it was not liberated into the cytosol even after extended incubation. The same experiment but run at 4 °C showed only impregnation of the polymer into the membrane as in the absence of pyrenebutyrate (see ESI†).
Conclusions
These studies were undertaken to determine if the cationic polymers 1 and 2 would import large and small, negative and positively charged, proteins into cells. If protein import was observed, then it was also important to determine if the polymer would “let go” of the protein once inside the cell. The data described shows that both the quaternary amine- and the guanidine-containing polymers 1 and 2 mediated import into the cells at 37 °C (but not at 4 °C). Once inside the cells, the quaternary amine polymer 1 releases more of the protein cargoes than its guanidine relative 2, irrespective of the charge and size of the proteins involved.
It was conveniently possible to determine the degree of endosomyl release for streptavidin Texas Red®. In this regard, it was again the quaternary amine polymer 1 that gave superior release (cf. less white regions indicating colocalization of all three labels in Fig. 3d as compared with Fig. 3h). Preliminary experiments indicate that the rate of import of proteins by these polymers is increased by pyrenebutyrate, just as Futaki et al. have observed in other systems.
Intracellular localization of the proteins featured in this study (GFP, β-galactosidase, and avidin) is of little interest. However, others focused on the delivery of more significant proteins might now consider import using these fluorescent polymers, and the quaternary amine 1 system in particular.
Experimental
Material and methods
See the ESI† for protocols and characterization information for polymers 1 and 2. Avidin-FITC and streptavidin-Texas Red® conjugates were purchased from Invitrogen by Life Technologies. β-Galactosidase was purchased from Calbiochem and labeled with Alexa Fluor 488®. GFP was kindly provided by Dr Wenshe Liu at Texas A&M University.
Cell culture
Clone 9 cells were cultured on 75 cm2 culture flasks in Ham’s Nutrient Mixture F-12 with 10% fetal bovine serum (FBS) in a humidified incubator at 37 °C with 5% CO2. Cells grown to subconfluence were plated in Lab-Tek two well chambered coverglass slides (Nunc) 2–3 d prior to the experiments.
CHO K1 cells were cultured on 75 cm2 culture flasks in DMEM supplemented with 10% FBS in a humidified incubator at 37 °C with 5% CO2. Again, cells grown to subconfluence were plated in Lab-Tek two well chambered coverglass slides 2–3 d prior to the experiments.
Fluorescence microscopy
Subcellular protein localization was measured on living Clone 9 and CHO K1 cells using a Zeiss 510 META NLO Multiphoton Microscope System consisting of an Axiovert 200 MOT microscope. Throughout, digital images were captured with a 40 ×/1.3 oil objective with the following filter sets:
for polymer 1 and 2: excitation 740 nm; emission BP 435–485
for FITC, Alexa Fluor® 488 and LysoTracker® Green DND-26 (abbreviated to “LysoTracker® Green” above): excitation 488 nm; emission BP 500–530
for the Texas Red® protein conjugates: excitation 543 nm; emission BP 565–615
Sequential optical sections (Z-stacks) from the basal-to-apical surfaces of the cell were acquired. Digital image acquisition was initiated approximately 1 μm below the basal surface of the cells and optical slices were collected at 0.5 μm steps through their apical surface using a high numerical objective lens (C-APO 63X/1.2 W CORR D = 0.28M27). These wide-field images were subjected to deconvolution using Intelligent Imaging Innovations (3I) software.
The protein : carrier complexes were pre-formed at room temperature for 1 h by mixing (mol : mol ratios throughout) the protein and the carrier in Ham’s Nutrient Mixture F-12 supplemented with 10% FBS for Clone 9 cells. The CHO K1 cells were supplemented with DMEM containing 10% FBS. Protein : carrier at 1 : 1 was used for avidin-FITC and streptavidin-Texas Red®, while 3 : 1 GFP, and 1.0 : 25 β-gal-Alexa Fluor® were used. To study the cellular uptake of the proteins, the culture medium was removed, the preformed protein : carrier complexes were added, and the cells were incubated for 12 h at 37 °C. After the incubation period, the cells were washed with phosphate-buffered saline (PBS, pH 7.4) and heparin solution several times before imaging.
Lysosomal colocalization
Clone 9 cells were incubated with streptavidin : polymer complex for 12 h at 37 °C. After the cells were washed with PBS and heparin, LysoTracker® Green was added and the cells were incubated for 30 min at 37 °C. The cells were washed again with PBS before imaging.
Cell viability assays
Clone 9 cells (3.5 K cells/well, 50 μL, in Ham’s medium) were plated on 96-well plates and allowed to adhere at 37 °C in 5% CO2 and 95% air for 3 h. The cells were then treated with 50 μL of each test compounds in PHFM-II (protein free medium) at 0 to 200 μM concentrations. The cells were then incubated for 72 h at 37 °C, then their viabilities were assessed through an MTT conversion assay.28 Briefly, 25 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide MTT (5 mg mL−1, in Hank’s balanced salt solution) were added and the cells were incubated for an additional 2 h. They were then lysed and the dark blue crystals solubilized with 100 μL of an aqueous solution containing 35% (v/v) N,N-dimethylformamide, 15% (v/v) glacial acetic acid, 15% (w/v) SDS with an adjusted pH of 3.8. The optical density of each well (at 570 nm) was measured with a BioTek Synergy 4 Microplate Reader. The viability of each cell line in response to the treatment with tested compounds was calculated as: % dead cells = 100 − (OD treated/OD control) × 100.
Supplementary Material
Acknowledgments
We thank The National Institutes of Health (GM087981), and The Robert A. Welch Foundation (A-1121) for financial support. We also thank Drs Aurore Loudet and Cliferson Thivierge for helpful discussions.
Footnotes
Electronic supplementary information (ESI) available: Synthesis of 1 and 2; delivery of streptavidin-Texas Red®, avidin-FITC, β-galactosidase-Alexa Fluor® 488 and GFP at 37 °C into Clone 9 and CHO K1 cells; delivery of BSA-Texas Red® at 4 °C mediated by 1; delivery of BSA-Texas Red® at 37 °C mediated by 1 in the presence of pyrenebutyrate as an additive; kinetic study of an effect of pyrenebutyrate on the rate of BSA-Texas Red® uptake at 37 °C. See DOI: 10.1039/c1ob05874f
Contributor Information
Robert C. Burghardt, Email: RBURGHARDT@cvm.tamu.edu.
Kevin Burgess, Email: burgess@tamu.edu.
References
- 1.McRae RL, Phillips RL, Kim IB, Bunz UHF, Fahrni CJ. J Am Chem Soc. 2008;130:7851–7853. doi: 10.1021/ja8007402. [DOI] [PubMed] [Google Scholar]
- 2.Feng X, Liu L, Wang S, Zhu D. Chem Soc Rev. 2010 [Google Scholar]
- 3.Pu KY, Li K, Shi J, Liu B. Chem Mater. 2009;21:3816–3822. [Google Scholar]
- 4.Feng X, Tang Y, Duan X, Liu L, Wang S. J Mater Chem. 2010;20:1312–1316. [Google Scholar]
- 5.Xu H, Gao SL, Yang Q, Pan D, Wang LH, Fan CH. ACS Appl Mater Interfaces. 2010;2:3211–3216. doi: 10.1021/am1006854. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Yang Q, Liu L, Wang S. Colloids Surf, B. 2011;85:8–11. doi: 10.1016/j.colsurfb.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Feng X, Lv F, Liu L, Tang H, Xing C, Yang Q, Wang S. ACS Appl Mater Interfaces. 2010;2:2429–2435. doi: 10.1021/am100435k. [DOI] [PubMed] [Google Scholar]
- 8.Magzoub M, Graeslund A. Q Rev Biophys. 2004;37:147–195. doi: 10.1017/s0033583505004014. [DOI] [PubMed] [Google Scholar]
- 9.Brooks H, Lebleu B, Vives E. Adv Drug Delivery Rev. 2005;57:559–577. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Morris MC, Depollier J, Mery J, Heitz F, Divita G. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 11.Saint PhD protein delivery kit http://www.Synvoluxproducts.com.
- 12.Bio Porter protein delivery kit http://www.sigmaaldrich.com.
- 13.Mahlum E, Mandal D, Halder C, Maran A, Yaszemski MJ, Jenkins RB, Bolander ME, Sarkar G. Anal Biochem. 2007;365:215–221. doi: 10.1016/j.ab.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Lee ALZ, Wang Y, Ye WH, Yoon HS, Chan SY, Yang YY. Biomaterials. 2008;29:1224–1232. doi: 10.1016/j.biomaterials.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 15.Vinogradov SV, Batrakova EV, Li S, Kabanov AV. J Drug Targeting. 2004;12:517–526. doi: 10.1080/10611860400011927. [DOI] [PubMed] [Google Scholar]
- 16.Didenko VV, Ngo H, Baskin DS. Anal Biochem. 2005;344:168–173. doi: 10.1016/j.ab.2005.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Futaki S. Adv Drug Delivery Rev. 2005;57:547–558. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Kosuge M, Takeuchi T, Nakase I, Jones AT, Futaki S. Bioconjugate Chem. 2008;19:656–664. doi: 10.1021/bc700289w. [DOI] [PubMed] [Google Scholar]
- 19.Loudet A, Han J, Barhoumi R, Pellois JP, Burghardt RC, Burgess K. Org Biomol Chem. 2008;6:4516–4522. doi: 10.1039/b809006h. [DOI] [PubMed] [Google Scholar]
- 20.Lee YJ, Erazo-Oliveras A, Pellois JP. ChemBioChem. 2010;11:325–330. doi: 10.1002/cbic.200900527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wender PA, Jessop TC, Pattabiraman K, Pelkey ET, VanDeusen CL. Org Lett. 2001;3:3229–3232. doi: 10.1021/ol0161108. [DOI] [PubMed] [Google Scholar]
- 22.Guterstam P, Madani F, Hirose H, Takeuchi T, Futaki S, El Andaloussi S, Graeslund A, Langel U. Biochim Biophys Acta, Biomembr. 2009;1788:2509–2517. doi: 10.1016/j.bbamem.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi T, Kosuge M, Tadokoro A, Sugiura Y, Nishi M, Kawata M, Sakai N, Matile S, Futaki S. ACS Chem Biol. 2006;1:299–303. doi: 10.1021/cb600127m. [DOI] [PubMed] [Google Scholar]
- 24.Guo ZS, Pei J, Zhou ZL, Zhao L, Gibson G, Lam S, Brug J. Polymer. 2009;50:4794–4800. [Google Scholar]
- 25.Wang H, Lu P, Wang B, Qiu S, Liu M, Hanif M, Cheng G, Liu S, Ma Y. Macromolecular Rapid Communications. 2007;28:1645–1650. [Google Scholar]
- 26.Sun B, Sun MJ, Gu Z, Shen QD, Jiang SJ, Xu Y, Wang Y. Macromolecules. 2010;43:10348–10354. [Google Scholar]
- 27.Perret F, Nishihara M, Takeuchi T, Futaki S, Lazar AN, Coleman AW, Sakai N, Matile S. Journal of the American Chemical Society. 2005;127:1114–1115. doi: 10.1021/ja043633c. [DOI] [PubMed] [Google Scholar]
- 28.Mosmann T. J Immunol Meth. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.