Figure 4.
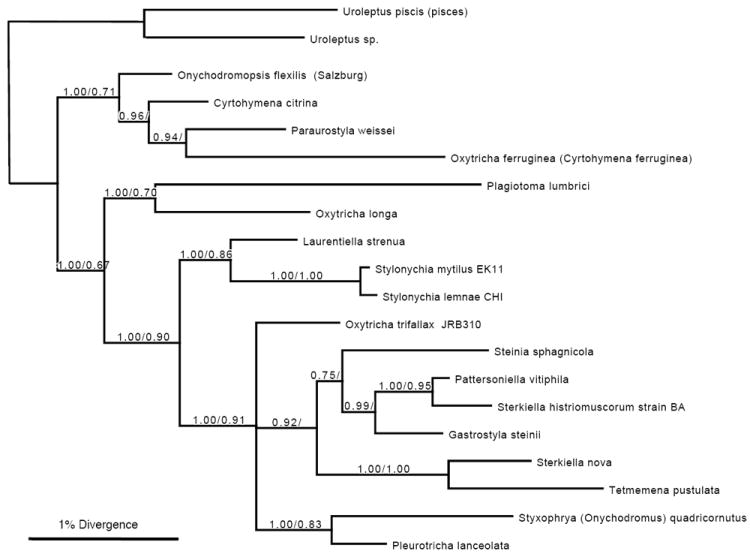
Bayesian and maximum likelihood phylogeny of small-subunit rRNA genes. Bootstrap values above 0.50 are shown (Bayesian/maximum likelihood). O. trifallax (JRB310), accession AF164121, is identical to O. trifallax AF508770; S. nova is accession X03948, S. lemnae CHI is AJ310496, S. mytilus EK11 is AJ310499, L. strenua AJ310487, S. sphagnicola AJ310494, P. vitiphila AJ310495, G. steinii AF508758, Tetmemena pustulata X03947 (formerly Stylonychia pustulata; identical to AF508775), Styxophya (also Onychodromus) quadricornutus X53485, P. lanceolata AF508768, O. longa AF508763, P. lumbrici AY547545, Oxytricha (also Cyrtohymena) ferruginea AF370027, P. weissei AJ310485, O. flexilis (Salzburg) AM412764, and C. citrina represented by AY498653. Uroleptus piscis (also pisces) AF508780, 98% identical to Uroleptus sp. AY294646 from in our lab (Chang et al. 2005), are both outgroups. S. histriomuscorum strain BA is new to this study with accession number HQ615720. The scale bar represents nucleotide divergence in Bayesian analysis.
