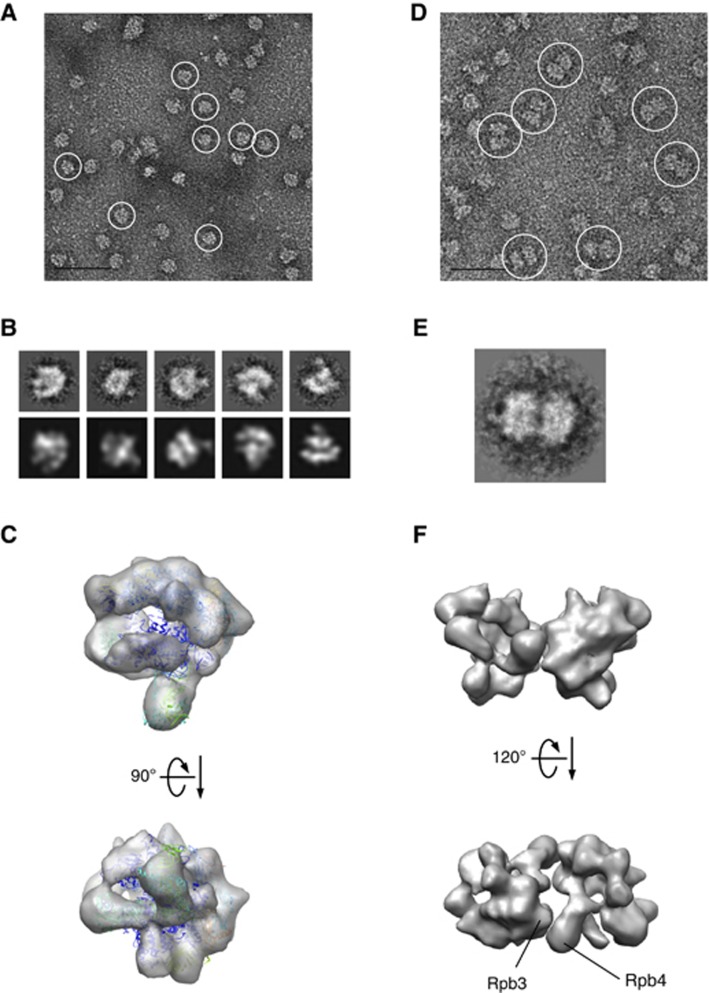
Figure 2.
Image analyses of native bovine RNAPII complexes. (A) Native bovine RNAPII(G), enriched in monomers (white circles), were preserved in negative stain (2% uranyl acetate, 50 nm bar scale). (B) Upper row: class averages of bovine RNAPII(G) particles obtained after reference-free alignment and clustering reveal renowned RNAPII features such as groove and stalk (Rpb4/7). Lower row: re-projections from the 3D reconstruction of native RNAPII(G) that best match the class averages above. (C) EM structure of the native bovine RNAPII(G) superimposed with the yeast RNAPII X-ray structure (PDB: 1WCM). (D) Native bovine RNAPII, enriched in dimers (white circles), preserved in negative stain (50 nm scale bar scale). (E) A representative class average of RNAPII dimer. (F) 3D model of the RNAPII dimer built from reconstruction RNAPII monomers. An orientation best matches the class average in (E) shows subunit Rpb3 and Rpb4 likely participate the dimer interface.