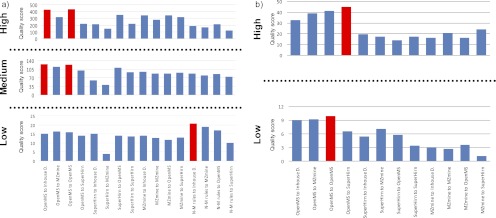
Fig. 6.
Comparison of the performance of 16 and 12 different combinations of feature detection/quantification and feature alignment/matching modules at high, medium, and low concentration differences of spiked peptides (see supplemental Tables S6 and S7) using the spiked human urine data set (a) and the spiked porcine CSF data set (b), respectively. Labels of the hybrid workflows (x axis) start with the name of the feature detection/quantification module followed by the name of the feature alignment/matching module. The best performing workflows at each concentration level difference are highlighted in red. The homogeneous OpenMS workflow and combinations of the OpenMS feature detection/quantification module with the in-house developed feature alignment/matching module result in the highest scores when concentration differences are large or medium for the spiked human urine data set (a), whereas the respective combination of the OpenMS-SuperHirn heterogeneous workflow provides the best performance for the porcine CSF data set spiked with large concentration differences (b). The scores level out at medium concentration differences, although some combinations do not perform well at any level (e.g. SuperHirn to MZmine). The combination N-M rules feature detection/matching module with the in-house developed feature alignment/matching module (Inhouse D.) performs best for low spiked concentration differences for spiked human urine data set (M-N rule peak picking was not performed for porcine CSF data set because of the incompatibility of this approach with high resolution data), whereas the best performing combination of feature detection/quantification and feature alignment/matching modules for the low spiked concentration difference of the porcine spiked CSF data set is the respective OpenMS homogenous workflow.