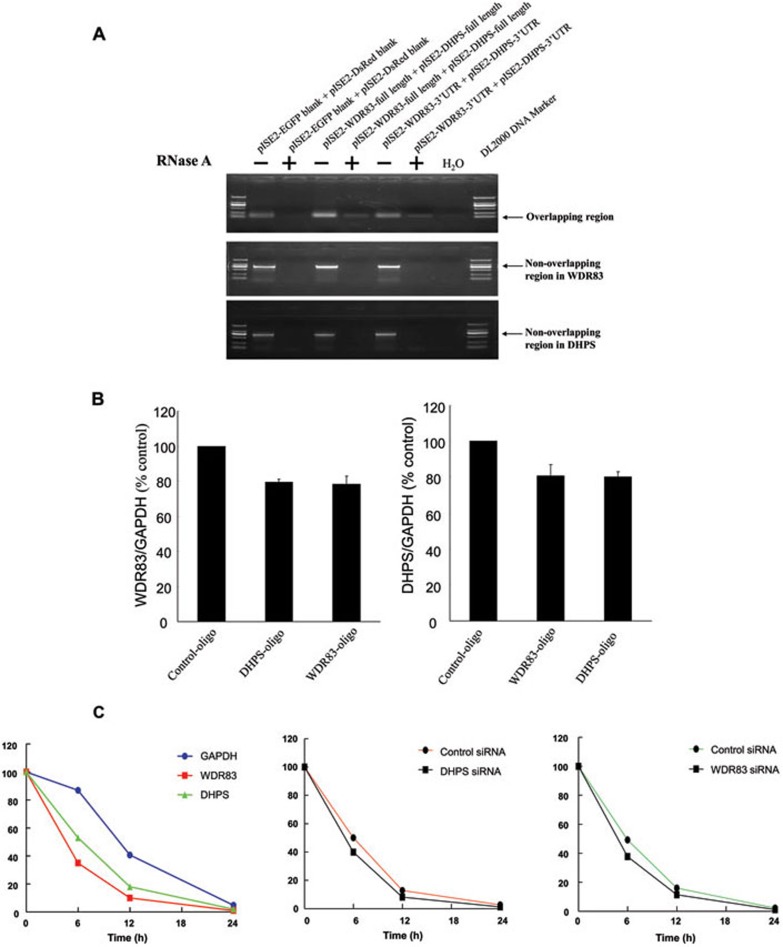
Figure 5.
DHPS and WDR83 formed RNA duplex and increased stability of each other. (A) RPA was performed on RNA samples from MGC803 cells. The plasmids of pIRES2-WDR83-full length and pIRES2-DHPS-full length were cotransfected in the same reaction, and the plasmids of pIRES2-WDR83-3′UTR and pIRES2-DHPS-3′UTR were also cotransfected in the same reaction. Total RNA was extracted and purified, single-stranded RNA was digested with RNase A, and the remaining double-stranded RNA was subjected to RT-PCR to amplify the overlapping or non-overlapping regions of WDR83 and DHPS. (B) Blocking the WDR83/DHPS RNA-RNA interaction with 2′-O-methyl oligoribonucleotides inhibited the bidirectional regulation between them. (C) Stability of WDR83 and DHPS over time was measured by qPCR relative to time 0 after blocking new RNA synthesis with α-amanitin (25 mM). MGC803 cells were transfected with siRNA against WDR83 or DHPS or control siRNA for 24 h, and were then further exposed to 25 mM α-amanitin for 6, 12 and 24 h. Cells were harvested and the stability of the WDR83 and DHPS mRNA was analyzed by qPCR. 18S RNA, which was a product of RNA polymerase I and was unchanged after α-amanitin treatment, was used as the control.