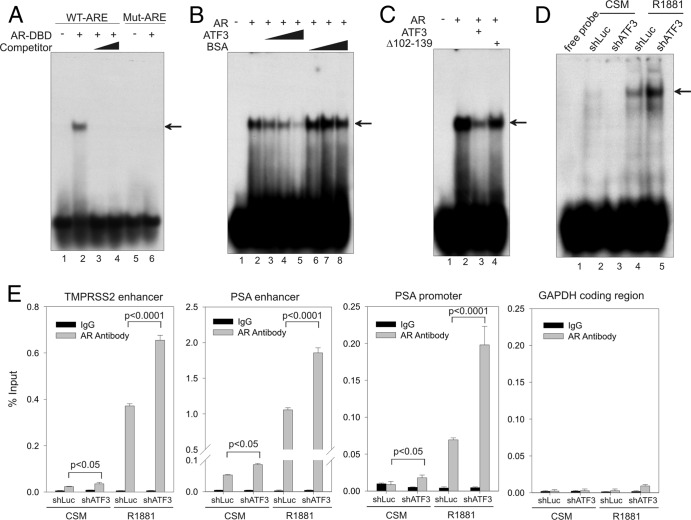
Fig 7.
ATF3 prevents AR from binding to ARE both in vitro and in vivo. (A) The purified AR-DBD protein (aa 537 to 644; 500 ng) was incubated with 32P-labeled oligonucleotide containing ARE derived from the PSA enhancer (lanes 1 to 4) or a mutated oligonucleotide (lanes 5 and 6) and subjected to gel shift assays. For competition assays (lanes 3 and 4), 50-fold and 100-fold amounts of unlabeled oligonucleotide were mixed with labeled oligonucleotide and AR-DBD protein. The arrow indicates the AR-ARE binding complex. (B) Increasing amounts of purified ATF3 proteins (100, 200, and 500 ng) or BSA were preincubated with AR-DBD protein at 4°C for 30 min and then subjected to gel shift assays. The arrow indicates the AR-DNA complex. (C) An equal amount (500 ng) of ATF3 or the ATF3 Δ102–139 mutant was preincubated with the AR-DBD protein for gel shift assays. The arrow indicates the AR-DNA complex. (D) LNCaP cells stably expressing shATF3 or shLuc were cultured in CSM for 2 days and then treated with 1 nM R1881 for 24 h. Nuclear extracts were prepared and incubated with 32P-labeled oligonucleotide as described for panel A. The main AR-DNA binding band is indicated by the arrow. (E) LNCaP cells expressing shATF3 or shLuc were treated as described for panel D and then fixed with formaldehyde for ChIP assays using anti-AR antibody or control IgG. Real-time PCR was used to quantify amounts of DNA fragments spanning the ARE in the TMPRSS2 enhancer, the PSA enhancer, or the PSA proximal promoter, as indicated. For specificity control, a random fragment in the GAPDH coding region was also amplified and quantified by real-time PCR. Data are depicted as averages ± standard deviations of three determinations. The P values were calculated using the Student t test.