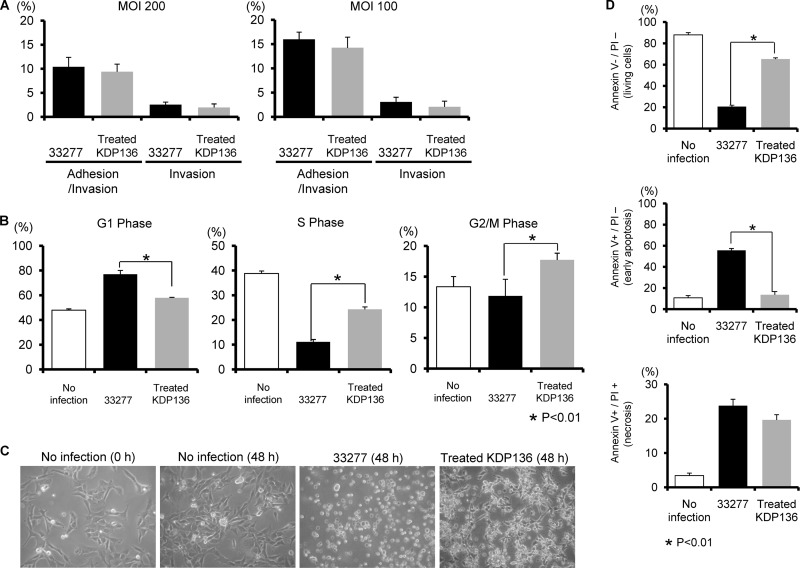
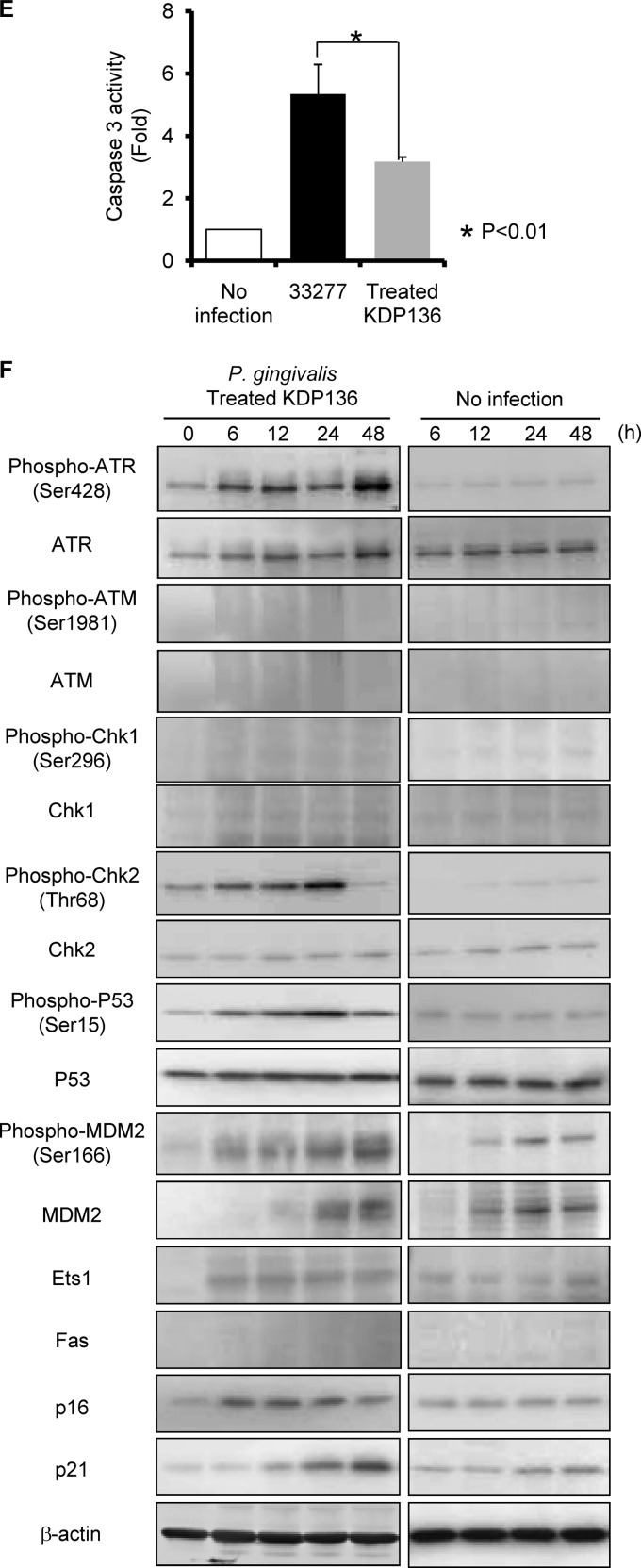
Fig 7.
HTR-8 cells infected with a P. gingivalis fimbriated gingipain-null mutant strain do not show G1 arrest and apoptosis. (A) Antibiotic protection invasion assay with P. gingivalis ATCC 33277 (wild type) and fimbriated KDP136 (fimbriated gingipain-null mutant strain). HTR-8 cells were infected with P. gingivalis strains at an MOI of 100 or 200 for 2 h. Numbers of adherent and/or intracellular bacteria were determined by viable counting of the cell lysates and are expressed as percentage of input bacterial cell number. Data are means ± SD of three independent experiments and were analyzed with a t test. (B) Cell cycle distribution was determined using flow cytometry. The fold change in numbers of cells in each phase of the cell cycle was calculated in comparison with noninfected cells. Data are means ± SD of three independent experiments and analyzed with a t test. (C) Light microscopy showing morphology of HTR-8 cells infected with P. gingivalis ATCC 33277 and fimbriated KDP136 at an MOI of 200 for 48 h. (D) Cells were stained with annexin V and PI and analyzed using flow cytometry. The fold change was calculated as described for panel B. The mean percentage ± SD of apoptotic (annexin V+/PI−) cells from three independent experiments was determined and analyzed with a t test. (E) Fold change in caspase 3 activity. Data are means ± SD from three independent experiments analyzed with a t test. (F) Cells were infected with P. gingivalis fimbriated KDP136 at an MOI of 200 for the indicated times. Lysates of infected and noninfected cells were subjected to immunoblotting to analyze expression profiles of DNA damage-responsive proteins, MDM2 phosphorylated at Ser166, MDM2, Ets1, Fas, p16, and p21. β-Actin was included as a loading control. Data are representative of three independent experiments.