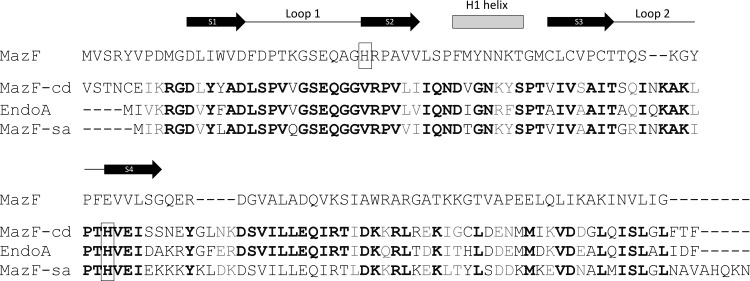
Fig 7.
MazF-cd exhibits significant sequence similarity to counterparts in S. aureus and B. subtilis. EndoA and MazF-sa were compared to each other and to E. coli MazF. Identical amino acids are highlighted in boldface, while conserved substitutions (+1 or greater on the BLOSUM62 alignment score matrix) are highlighted in gray. Histidines believed to be involved in sequence recognition are boxed. Only the secondary structure motifs relevant to this work are illustrated above the respective amino acids of E. coli MazF. Black arrows, β-sheets; gray rectangle, α-helix; black line, unstructured loops.