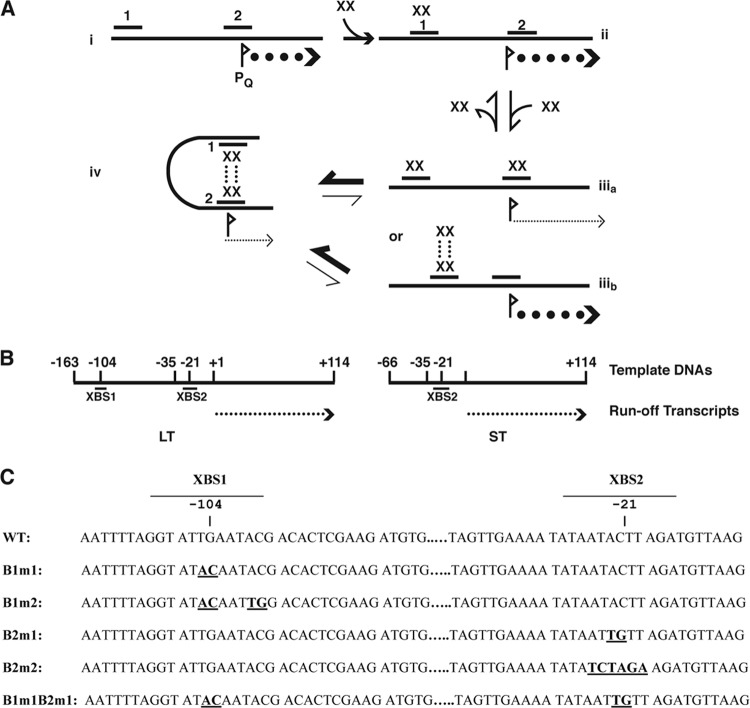
Fig 1.
Regulatory circuits controlling the initiation of the E. faecalis pheromone response in the pCF10 system. (A) Potential states of occupancy of the PrgX DNA binding sites (XBSs) in the prgQ promoter region. (i) No PrgX present and maximum expression of prgQ. (ii) For a DNA target containing both binding sites, at low PrgX concentrations, dimers bind first to the higher-affinity site (>100× higher than the affinity of PrgX for XBS2) XBS1, but occupancy of this site has no direct effect on PQ. (iiia) Occupancy of both XBSs is predicted to reduce prgQ transcription by steric hindrance of RNA polymerase binding, but affinity for XBS2 is <1/100th that of the affinity for XBS1 (3). (iiib) Since PrgX tetramers have been observed in crystals (20, 27), it is theoretically possible to have XBS1 bound by a tetramer. (iv) Protein-protein interactions between pairs of PrgX dimers bound to the two XBSs results in a DNA loop that increases the stability of the repressing complex. Data presented in this paper and elsewhere (3) suggest that the affinity of the PrgX dimer for XBS2 is sufficiently low in the absence of DNA looping, that conversion between states ii and iv occurs rapidly, and states iiia and iiib, or a structure where only XBS2 is bound, probably do not exist at sufficient levels to be biologically important. Pheromone (cCF10) and inhibitor (iCF10) peptides do not directly alter the DNA binding regions of PrgX; instead they appear to affect the PrgX oligomerization state, with iCF10 predicted to drive the system toward state iv, with cCF10 having the opposite effect (20, 27). (B) Linear templates for in vitro transcription. The positions of the ends of the templates relative to the “+1” nucleotide of prgQ mRNA are indicated along with the locations of the XBSs and PQ regions. The long template (LT) contains both XBSs, whereas the short template (ST) contains only XBS2. The runoff transcript in both cases is 114 nt. (C) Sequences of the XBS regions of pCF10. Mutations used in this study are shown in boldface, and the sequence coordinates relative to the experimentally determined “+1” nucleotide of the prgQ transcript are indicated. WT, wild type.