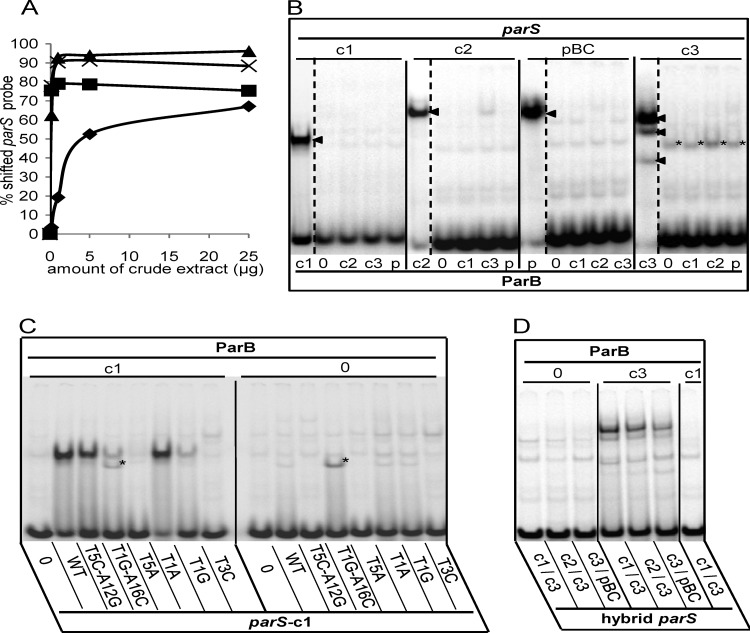
Fig 2.
EMSA of the Par systems of B. cenocepacia. (A) Binding of each B. cenocepacia parS probe was tested with increasing amounts (0.2, 1, 5, and 25 μg) of crude extracts of E. coli cells overproducing its cognate ParB (▲, c3; ×, pBC; ■, c2; ◆, c1). (B) Radiolabeled parS probes, denoted above each group of lanes, were incubated with 10 mg of crude extract protein containing ParB-c1 (lanes c1), -c2 (lanes c2), -c3 (lanes c3), or -pBC (lanes p) or no ParB (lanes 0). The cognate ParB-parS interactions produce a single shifted complex in the case of c1, c2, and pBC and a three-band pattern in the case of c3 (arrowheads). The parS-c3 fragment is also retarded by an unknown E. coli protein (*). (C) Protein extracts containing ParB-c1 or no ParB, indicated by “c1” and “0” above the lane groups, were incubated with the radiolabeled 26-bp probes indicated below each lane as follows: negative-control sequence CTAGTCGTACGACTAG (lanes 0), wild-type parS-c1 (lanes WT), and parS-c1 mutated as indicated (other lanes). The mutated parS-c1 T1G-A16C fragment is shifted by an unknown E. coli protein (*). (D) Cell extracts containing no ParB, ParB-c3, or ParB-c1 (as indicated) were incubated with the hybrids indicated below each lane.