Fig 6.
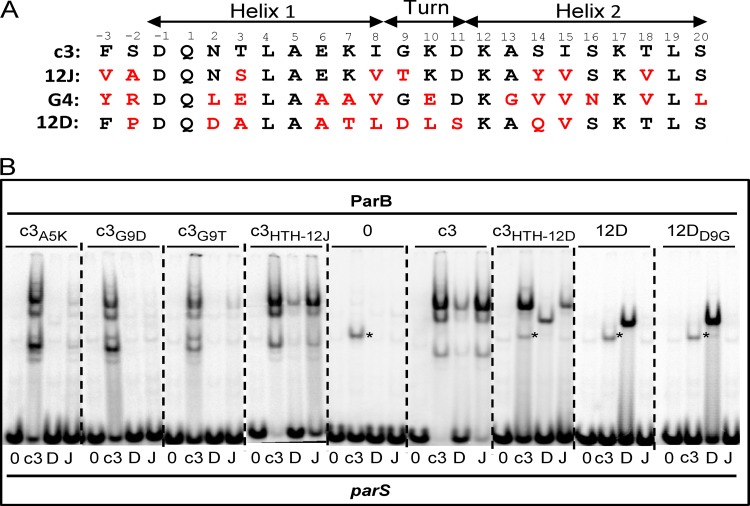
Modifications in a putative HTH domain of ParB-Bsr3. (A) Comparison of the putative HTH domains of ParB-c3, -12J, -G4, and -12D. Amino acids identical to ParB-c3 are in black, others in red. Positions are numbered and presumed helices are shown. (B) EMSA of ParB proteins with the HTH domain exchanged (from −3 to 20 according to the numbering in panel A) or modified as shown above each panel, e.g., for c3A5K, ParB-c3 with Lys substituted for Ala at position 5. The parS probes are denoted below each lane as follows: 0, negative-control sequence CTAGTCGTACGACTAG; c3, parS-c3; D, parS Ralstonia pickettii 12D plasmid 1; J, main parS Ralstonia pickettii 12J. The parS-c3 fragment is also retarded by an unknown E. coli protein (*). Note that the three-band pattern typical of ParB-c3, possibly resulting from protein cleavage at sites distant from the HTH, is replaced by a single band after substitution with the 12D HTH. This exchange would not be expected to eliminate protease sensitivity of two sites, suggesting that the three-band pattern could have another explanation, such as formation by ParB-c3 of three types of complex with parS sites, each with a different gel electrophoretic mobility.