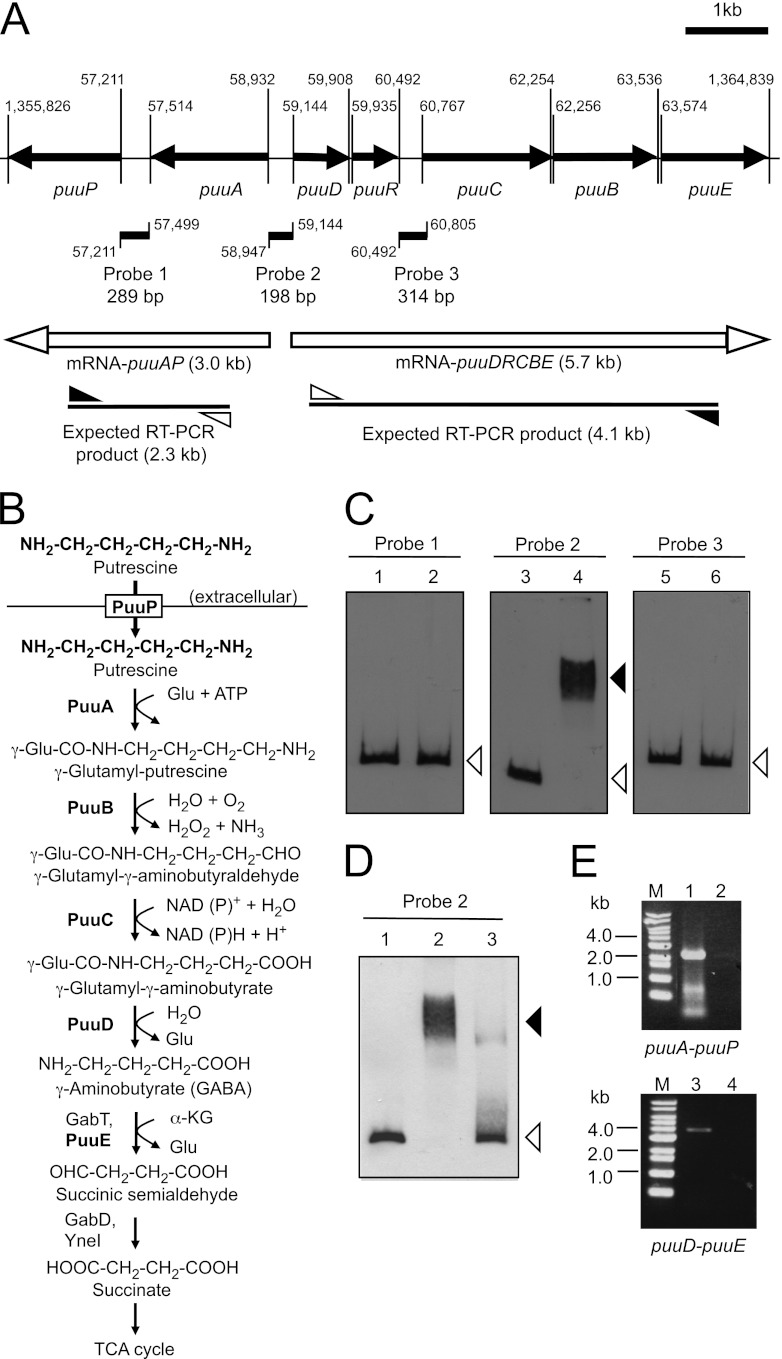
Fig 1.
Gel mobility shift assay to detect PuuR and DNA probe complexes. (A) Depiction of the puu gene cluster and locations of probes, which were designed for gel mobility shift assay at the intergenic regions between puuP and puuA (probe 1), puuA and puuD (probe 2), and puuR and puuC (probe 3). puuR and other puu genes are shown as black arrows with the gene names below. The locations of the three probes are indicated by black lines annotated with the probe names and lengths. Numbers indicate the start and end bases of the probes and open reading frames in the E. coli genome. The numbers omit the first two digits, except for the ends of puuP and puuE. mRNAs expressed from puuA and puuD promoters are labeled with white arrows and the estimated sizes of the transcripts. The expected RT-PCR products from mRNA-puuAP and mRNA-puuDRCBE are labeled with lines and the estimated sizes. White and black triangles indicate forward (puuA-R for mRNA puuAP and puuD-RT1 for mRNA-puuDRCBE) and reverse (puuP-RT1 for mRNA-puuAP and puuE-RT2 for mRNA-puuDRCBE) primers, respectively. (B) The Puu pathway of E. coli. PuuP works as a putrescine importer from extracellular space. PuuA, PuuB, PuuC, and PuuD make up the Puu pathway that degrades putrescine to GABA via γ-glutamylated intermediates. Glu, glutamic acid. α-KG, α-ketoglutarate. (C) Gel mobility shift assay using three probes and His6-PuuR. Assays were conducted using 5% PAGE in 0.5× TBE buffer. Thirty femtomoles (final concentration, 3 nM) of DIG-labeled probe 1, probe 2, and probe 3 was incubated at 4°C for 20 min in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 40 pmol (final concentration, 4 μM) of purified His6-PuuR. Black and white triangles indicate signals of the shifted band and free probe, respectively. (D) Gel mobility shift assay using probe 2 and cell extract from KEI23 (His6-PuuR) and GM13 (His6-PuuR3Ala). Cell extracts were prepared from the cells after 5 h of IPTG induction. For each assay, 3.6 μg cell extract protein was mixed with 30 fmol of DIG-labeled probe 2. Lane 1, absence of cell extract. Lane 2, presence of cell extract with His6-PuuR. Lane 3, presence of cell extract with His6-PuuR3Ala. Black and white triangles indicate signals of the shifted band and free probe, respectively. (E) Transcription unit of the puu genes identified by RT-PCR and agarose gel electrophoresis of RT-PCR products. Total RNA was prepared from strain KEI12, and cDNA was synthesized by an RT reaction using primer puuP-RT1 or puuE-RT2, respectively. PCR was then performed using the primer sets described above with the cDNA as the template. Control reactions without RT enzyme were performed by the same method. Lanes contain the following samples: M, 1-kb DNA ladder (Bioneer); 1 and 2, RT-PCR product and control sample without RT using the puuA-R and puuP-RT1 primer set; 3 and 4, RT-PCR product and control sample without RT using the puuD-RT1 and puuE-RT2 primer set.