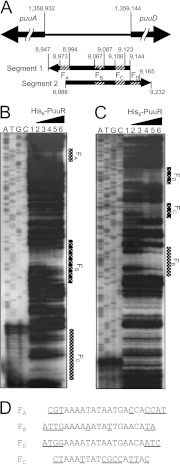
Fig 2.
DNase I footprint analysis of the intergenic region between puuA and puuD. (A) Location of two [32P]ATP-labeled DNA segments between puuA and puuD. Arrows indicate directions from 5′ to 3′ on 32P-labeled DNA strands. Striped areas labeled FA, FB, FC, and FD indicate PuuR binding regions, which were identified by DNase I footprint analysis. Numbers indicate the start and end bases of the segment and the PuuR binding region in the E. coli genome. The numbers omit the first three digits, except for the start of puuA and puuD. (B and C) DNase I footprint assay with His6-PuuR. The 32P-labeled probes (1.35 fmol) were incubated in the absence (lane 1) or presence of increasing concentrations of purified His6-PuuR (lane 2, 1 pmol; lane 3, 5 pmol; lane 4, 10 pmol; lane 5, 20 pmol; lane 6, 40 pmol), and then subjected to DNase I footprinting assays (panel B represents segment 1 from panel A; panel C represents segment 2 from panel A). Lanes A, T, G, and C represent the respective sequence ladders. (D) An alignment of the nucleotide sequences of all four sites protected by PuuR, which were observed in the DNase I footprint assay. Bases in the predicted 15-bp PuuR recognition sequence (AAAATATAATGAACA) are shown, and bases not in the predicted consensus sequence are shown with underlined letters.