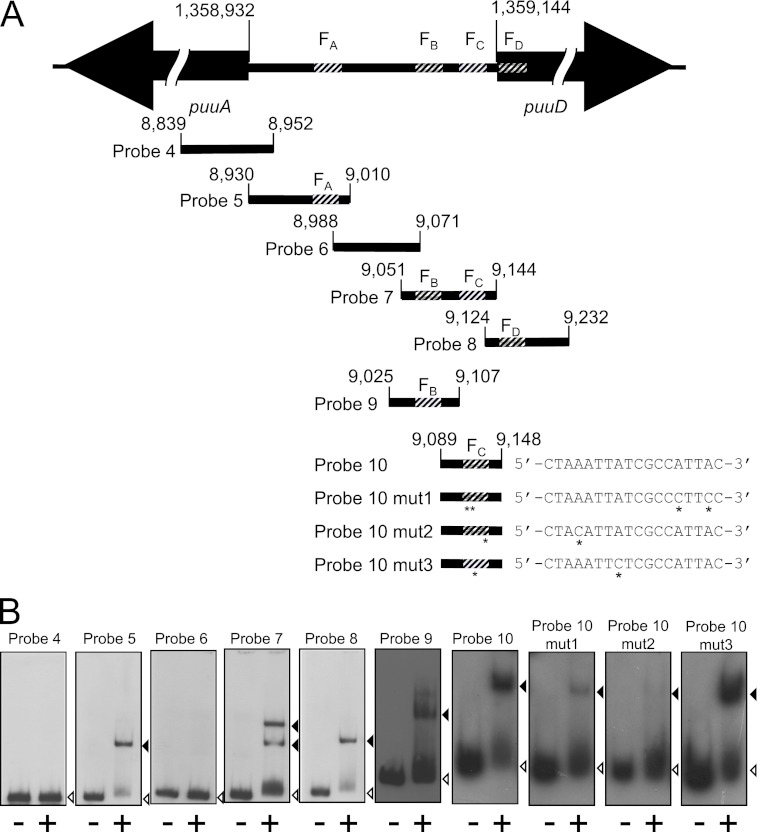
Fig 3.
Gel mobility shift assays using probes designed from the intergenic region of puuA and puuD. (A) Intergenic region between puuA and puuD, and location of each probe used in gel mobility shift assays. The base number of the 5′ end of puuA and puuD in the E. coli genome is indicated above each gene. The four PuuR binding sites, FA, FB, FC, and FD, are shown as striped areas. The probe location is indicated with the base numbers in the E. coli genome. The number omits the first three digits except for the start of puuA and puuD. Sequence alignments of each FC site mutation are shown. Asterisks below probes 10 mut1, 10 mut2, and 10 mut3 and sequences indicate mutation sites, which were introduced using the QuikChange method in the FC site. (B) Gel mobility shift assays were performed following the same method as described for Fig. 1 except for the concentration of the probes. Probe 9, probe 10, probe 10 mut1, probe 10 mut2, and probe 10 mut3 were prepared at 1.5 pmol/μl, and 1 μl of each probe was used in the reactions. Lanes on the left (marked with minus symbols below each lane) were incubated in the absence of His6-PuuR, and lanes on the right (marked with a plus symbol below each lane) were incubated in the presence of His6-PuuR. Black and white triangles indicate signals of the shifted band and free probe, respectively.