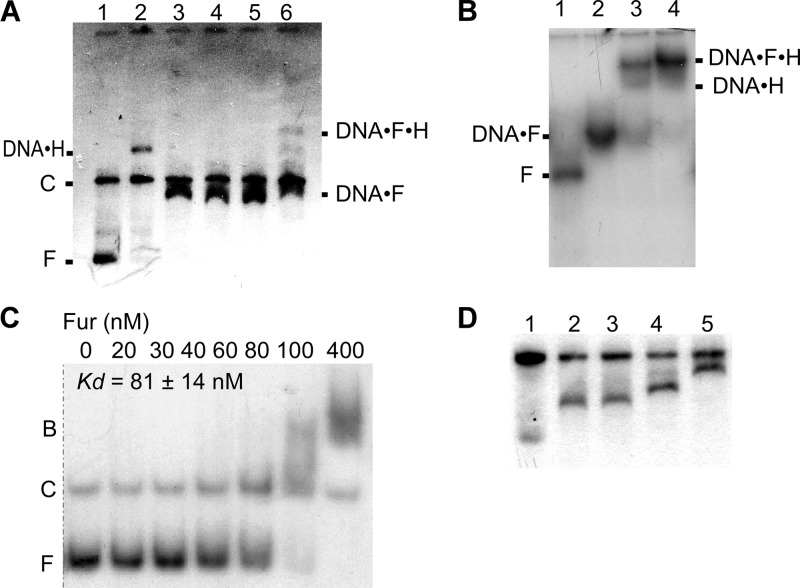
Fig 6.
Electrophoresis mobility shift analysis of Fur-hlyII DNA interaction. (A) EMSA of Fur and HlyIIR binding to PhlyII. Plasmid pUJ1 was digested with EcoRI and BamHI. Lanes: 1, no protein added; 2, HlyIIR at 0.1 μM; 3 to 5, Fur at 0.4, 0.8, and 1.2 μM, respectively; 6, Fur at 0.8 μM after 5 min incubation (25°C) with 0.1 μM HlyIIR added. Positions of free (F) and competitor (C) DNA and DNA-protein complexes are shown. (B) HlyIIR and Fur bind to a 400-bp DNA fragment of the entire hlyII promoter-operator. Lanes: 1, no protein added; 2, Fur at 0.8 μM; 3, Fur at 0.8 μM and HlyIIR at 0.2 μM; 4, Fur at 0.8 μM and HlyIIR at 0.4 μM. (C) Determination of the apparent equilibrium binding constant (KD) of Fur-PhlyII interactions by EMSA. 5′-end-labeled DNA fragments (20 nM) were incubated in transcription buffer with the concentration of Fur shown below each lane. The samples were separated on a 6% polyacrylamide gel. Positions of bound (B) and free (F) DNA are shown. (D) Resolution of Fur-PhlyII complexes with different stoichiometries. DNA was processed as for Fig. 4A. Lanes: 1, no protein, with doubled DNA to ensure visibility of diffuse small fragments; 2, Fur at 0.3 μM; 3, Fur at 0.6 μM; 4, Fur at 1.2 μM; 5, Fur at 2 μM. Experiments for panels A, B, and D were done using 0.7% agarose.