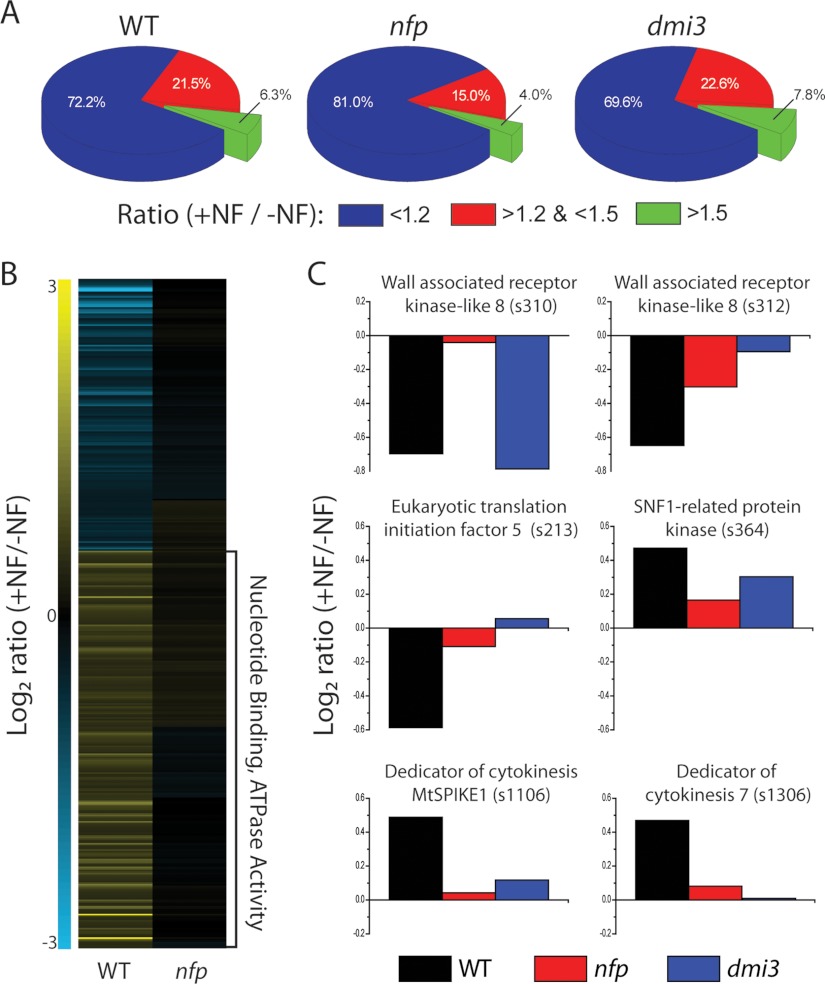
Fig. 3.
Global view of changes in the proteome and phosphoproteome from all the experiments combined. A, Pie charts displaying the distribution of phosphorylation changes for wild-type (WT), nfp and dmi3. Wild-type plants readily response to NF as 6.3% of phosphoisoform are altered more than 1.5-fold. Nfp displayed a lower response, 4.0%, but the presence of these changes provides evidence of a separate NF receptor sensing NF. Dmi3 more NF responses than wild-type plants, as the phosphorylation state of 7.8% of phosphoisoforms was altered more than 1.5-fold. B, Heatmap of phosphoisoforms altered in wild-type and not in nfp. Phophoisofroms exhibiting a fold change more than 1.35 in wild-type plants and less then 1.25 in nfp were grouped via hierarchical clustering. Functional analysis revealed phosphoisoforms, which demonstrate an up-regulation in phosphorylation upon NF treatment are enriched for the terms nucleotide binding and ATPase activity. C, Representative proteins significantly altered in wild-type and not in nfp. Six pohsphoisoforms that were significantly (p < 0.05, Student's t test, assuming equal variances) altered more than 1.35-fold in wild-type plants and less than 1.25-fold in nfp demonstrate the use of comparing wild-type and mutant measurements. These examples contain phosphoisoforms, which display both nfp dependent regulation and dmi3 dependent regulation.