Figure 2. Deletion analysis of TR reveals DNA sequence required for mLANA-mediated repression.
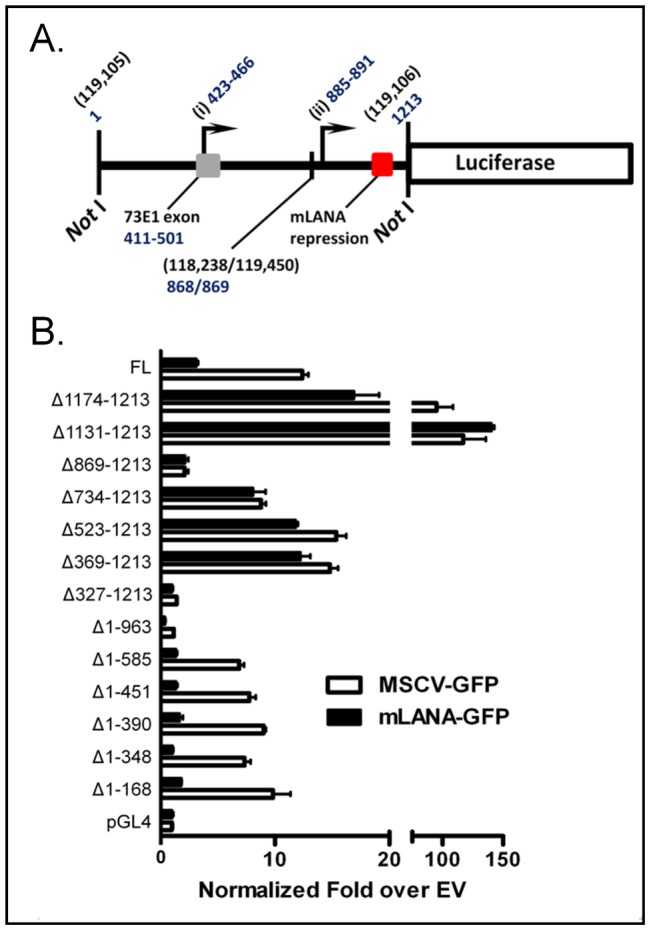
(A) Schematic of pGL-TR, noting the numbering scheme with regard to the NotI site. Coordinate 1 is corresponds to MHV68 WUMS [31] bp 119,105, proceeding in order towards the right end of the unique sequence. Coordinate 894 is the end of one TR unit fused to the beginning of the next. Indicated on the diagram are two known sites of transcription initiation: (i) (73p1) starts transcription between coordinates 423–466 (118,683–118,640) depending on cell type [29], [32] and the partial exon splices to the full 73E1 exon (118,695–118,605) in the next repeat unit; (ii) initiates transcription of one species of ORF75a beginning between coordinates 885–891 (118221–118216) [32] (promoter elements are likely shared between ORF75a and ORF73 transcripts that initiate at 73E2 [29], and in distal copies may splice into 73E1). (B) Serial deletions of the TR were made in the pGL-TR vector by PCR and are named accordingly. Each TR deletion, including the full-length TR (FL) was co-transfected into 293T cells, along with either an mLANA-GFP or empty GFP expression vector, as in Fig. 1, in triplicate. Here, data are normalized by taking the ratio of each deletion construct over its respective pGL4.10 empty reporter control, setting pGL4.10 in each case to 1. Deleting the 3′ end gives a large uptick in activity, which is still repressed by mLANA, until the second deletion removes mLANA sensitivity. 5′ deletions had no effect on mLANA sensitivity.
