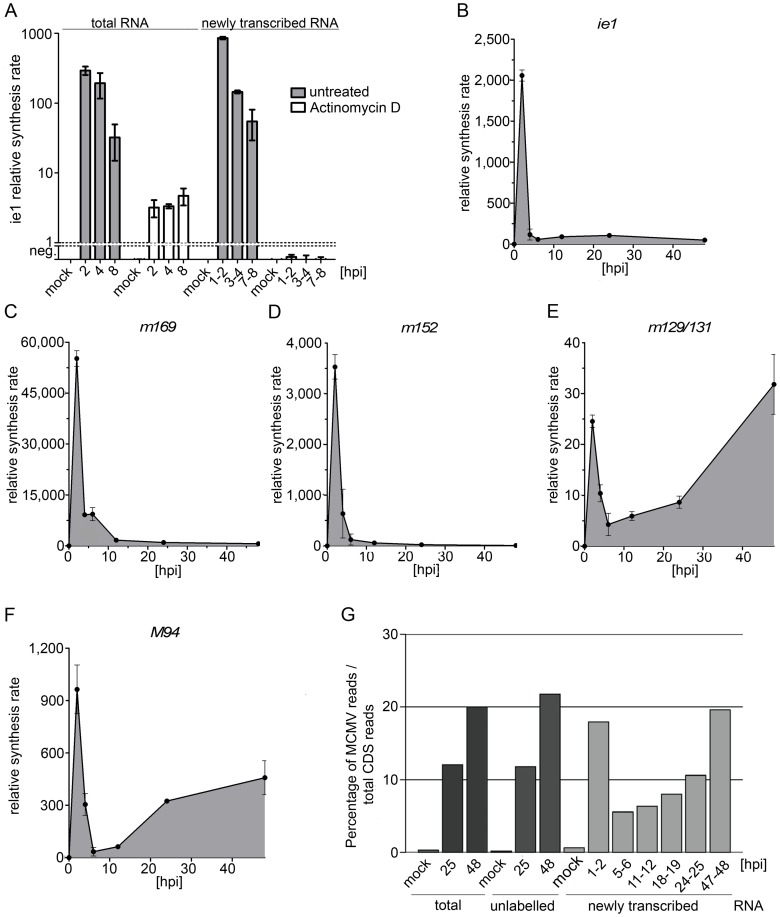
Figure 5. Real-time kinetics of viral gene expression.
(A) 4sU-tagging allows efficient removal of viral DNA and virion-associated RNA. Newly transcribed RNA was labeled with 200 µM 4sU for 1 h in MCMV-infected NIH-3T3 cells at −1 to 0 (mock), 1–2, 3–4 and 7–8 hpi. As a negative control, Actinomycin-D was added to cells prior to infection to block transcription and thus 4sU-incorporation. Total RNA was isolated, treated with DNaseI and newly transcribed RNA was purified. qRT-PCR analysis was performed on newly transcribed RNA for viral ie1 and cellular Lbr. Shown are the combined data (means +/− SD) of three independent experiments. (B–F) Gene expression kinetics of exemplary viral genes. Shown are qRT-PCR measurements of newly transcribed RNA for ie1 (B), the early genes m169 (C) and m152 (D) as well as for the late genes m129/131 (E) and M94 (F). Synthesis rates were normalized to Lbr expression. Shown are the combined data (means +/− SD) of three independent experiments. (G) Contribution of viral transcripts to all coding sequence reads (CDS). RNA-seq was performed on newly transcribed, total and unlabeled pre-existing RNA samples (n = 1). Reads were mapped to both the cellular and viral transcriptome/genome. The contribution of viral reads to all CDS reads at different times of infection is shown.