Figure 5. Experimental strategy to test rebound DNA methylation after Dicer1 ablation.
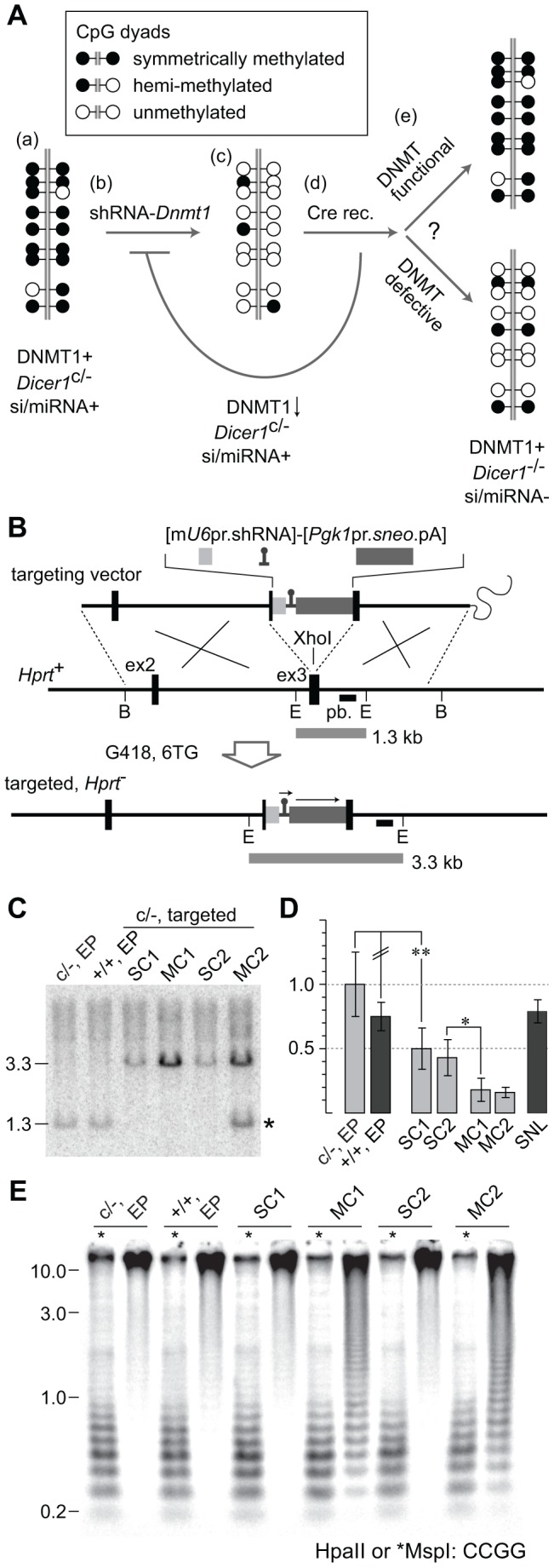
(A) (a) The parental Dicer1 c/− XY ES cell line is normally methylated. (b) A construct containing an shRNA targeting Dnmt1 mRNA (shRNA-Dnmt1) is homologously recombined into the X-linked Hprt locus. (c) The integrated shRNA effects substantial reduction in genome-wide DNA methylation content, although a background of DNA methylation is retained due to ongoing de novo DNMT activity. (d) Recombinant clones are transiently transfected with Cre to ablate the Dicer1 c allele. After ablation, clearing of residual DICER1 and si- and miRNAs, as well as restoration of functional DNMT1 activity, is expected to occur within a few cell divisions. (e) DNA methylation would potentially be re-established over ∼5 passages, corresponding to ∼25 cell divisions through the combined action of the DNMT system as a whole. The extent of rebound DNA methylation serves as an indicator of the dependency of this process on DICER1 activity and the presence of si- and miRNAs. (B) The targeting vector consisted of a 6.8 kb BamHI (B) genomic fragment of Hprt spanning exons 2 and 3 (ex2 and ex3). The mU6 promoter (pr) driving the shRNA and a sneo selection cassette were inserted into ex3. The sequence of the shRNA cassette is provided (Text S1). All clones surviving G418 and 6TG dual selection are expected to be targeted. pb, probe used in Southern blot, hybridizing to EcoRI (E) bands as indicated by horizontal bars. (C) Southern blot to detect gene targeting. WT bands are seen in the parental Dicer1 c/− line, and in a Dicer1 +/+ line. An increase in band size is occurs when the single Hprt sequence is replaced by a single copy (SC) or multiple copy concatemer (MC) of the targeting vector. In addition to the mutant band, clone MC2 clone shows a WT band of normal intensity (*). This results from recombination occurring in the 5′ arm, rather than in the 3′ arm, of the terminal vector in the concatemer. (D) qPCR of Dnmt1 mRNA. All values were calibrated to that obtained for the Dicer1 c/− line, adjusted to 1.0. SNL, mitomycin C-inactivated feeder cells. Other details as above. (E) Southern blot to detect methylation levels at MinS repeats. Details as above.
