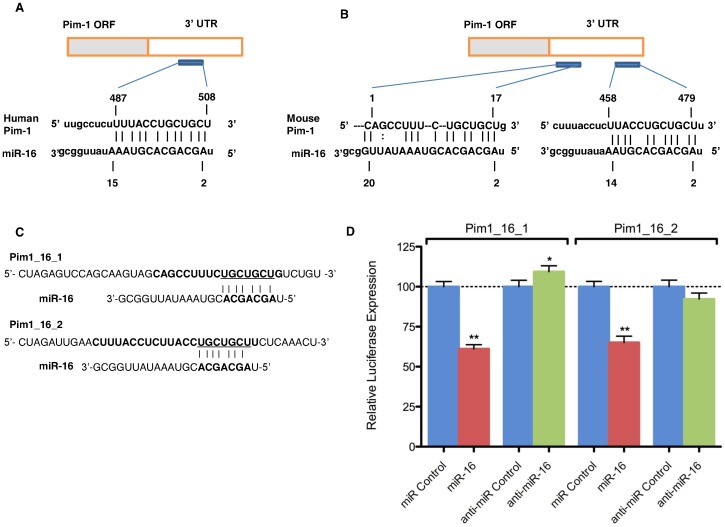
Figure 5. Pim-1 is a direct target of miR-16.
(A) Schematic representation of the human Pim-1 transcript. Predicted miR-16 binding site is depicted. The numbers (+487–508) represent the nucleotides (relative to Pim-1 termination codon) that are predicted to base pair with the miR-16 seed sequence. (B) Schemateic representation of the mouse Pim-1 transcript with predicted miR-16 binding site. The numbers are represented like in panel A. (C) Schematic representation of luciferase constructs used for reporter assays. Pim1_16_1 was designed on the basis of +1–17 region of Pim-1 3′UTR and Pim1_16_2 and was designed on the basis of +458–479 region of Pim-1 3′UTR. (D) Luciferase assays in HEK-293 cells transfected with vectors shown in panel C, miR-16 and control oligonucleotides. Bars represent luciferase activity for the corresponding vectors.**, p<0.01; *, p<0.05.