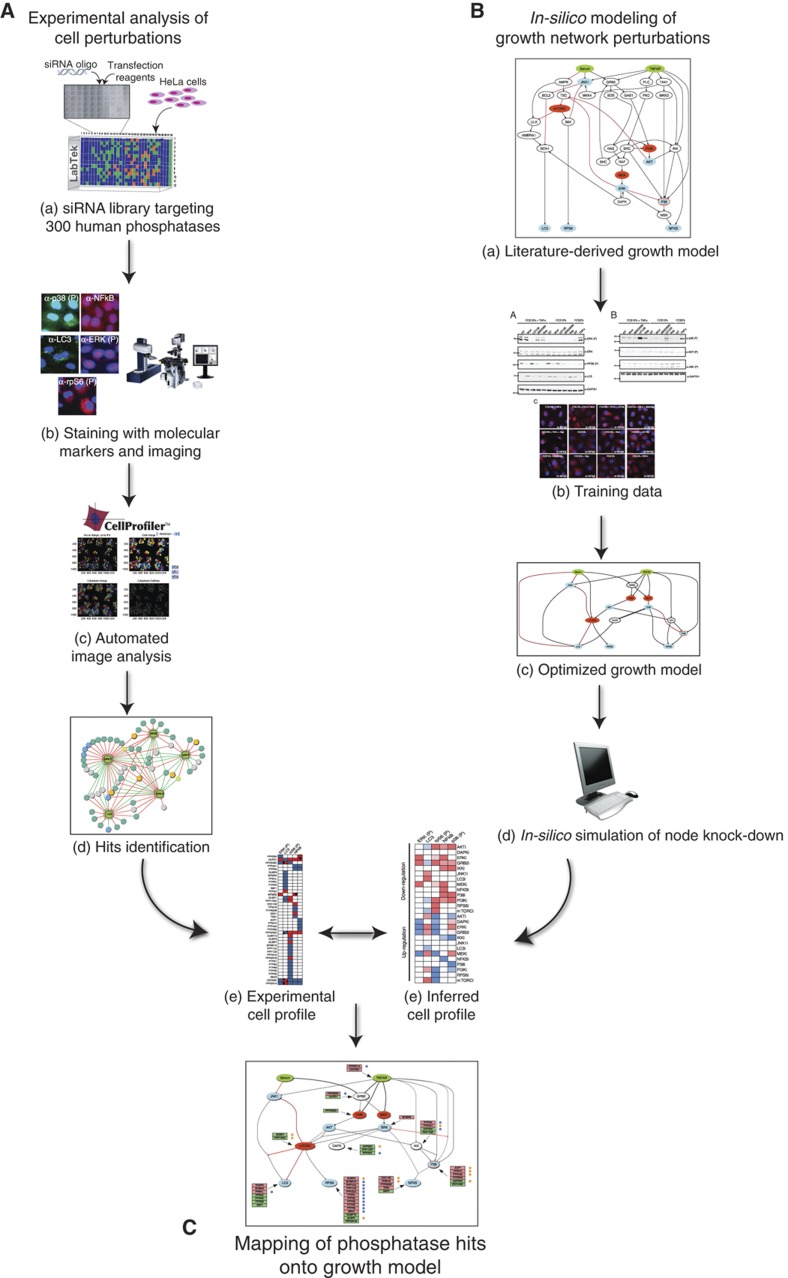
Figure 1.
Schematic illustration of a strategy to map protein phosphatases onto growth pathways. (A) We used a multiparametric siRNA phenotypic screening of the set of human genes encoding protein containing phosphatase domains or regulatory subunits to identify, by automated fluorescence microscopy and automated image analysis, genes whose downregulation modulates the activity of some key growth-associated pathways. The perturbation of the cell state by each siRNA is represented as a vector whose coordinates are the measured readouts. (B) In parallel, we assembled a literature-derived signed directed network and we simplified and optimized it by training with experimental data. The resulting logic growth model was used to infer the cell state upon perturbation of each node. (C) Finally, by matching the experimentally determined cell states with the one predicted by the pathway model, we inferred the pathway nodes that were likely to be affected by the phosphatase knock-down.